Abstract
Clostridium sp. strain TO-931 can rapidly convert the primary bile acid cholic acid to a potentially toxic compound, deoxycholic acid. Mixed oligonucleotide probes were used to isolate a gene fragment encoding a putative bile acid transporter from Clostridium sp. strain TO-931. This DNA fragment had 60% nucleotide sequence identity to a known bile acid transporter gene from Eubacterium sp. strain VPI 12708, another bile acid-7α-dehydroxylating intestinal bacterium. The DNA (9.15 kb) surrounding the transporter gene was cloned from Clostridium sp. strain TO-931 and sequenced. Within this larger DNA fragment was a 7.9-kb region, containing six successive open reading frames (ORFs), that was encoded by a single 8.1-kb transcript, as determined by Northern blot analysis. The gene arrangement and DNA sequence of the Clostridium sp. strain TO-931 operon are similar to those of a Eubacterium sp. strain VPI 12708 bile acid-inducible operon containing nine ORFs. Several genes in the Eubacterium sp. strain VPI 12708 operon have been shown to encode products required for bile acid 7α-dehydroxylation. In Clostridium sp. strain TO-931, genes potentially encoding bile acid-coenzyme A (CoA) ligase, 3α-hydroxysteroid dehydrogenase, bile acid 7α-dehydratase, bile acid-CoA hydrolase, and a bile acid transporter were similar in size and exhibited amino acid homology to similar gene products from Eubacterium sp. strain VPI 12708 (encoded by baiB, baiA, baiE, baiF, and baiG, respectively). However, no genes similar to Eubacterium sp. strain VPI 12708 biaH or baiI were found in the Clostridium sp. strain TO-931 bai operon, and the two putative Eubacterium sp. strain VPI 12708 genes, baiC and baiD, were arranged in one continuous ORF in Clostridium sp. strain TO-931. Intergene regions showed no significant DNA sequence similarity, but primer extension analysis identified a region 115 bp upstream from the first ORF that exhibited 58% identity to a bai operator/promoter region identified in Eubacterium sp. strain VPI 12708. These results indicate that the gene organization, gene product amino acid sequences, and promoters of the bile acid-inducible operons of Clostridium sp. strain TO-931 and Eubacterium sp. strain VPI 12708 are highly conserved.
In mammals, the primary bile acids cholic acid and chenodeoxycholic acid are synthesized in the liver and conjugated to either glycine or taurine (31). Conjugated bile acids are required for the proper digestion and absorption of cholesterol, lipids, and other lipid-soluble compounds. Bile acids are actively absorbed in the terminal ileum and returned to the liver (15). However, some bile acids pass into the large intestine and are extensively biotransformed (4). In particular, a minute population of bacteria can 7α-dehydroxylate the primary bile acids into secondary bile acids, generating potentially toxic products. The bile acid 7α-dehydroxylation products of cholic acid and chenodeoxycholic acid are deoxycholic acid and lithocholic acid, respectively (4, 15).
In humans, increased levels of deoxycholic acid in the bile acid pool have been associated with an increased risk of cholesterol gallstone disease (6, 9, 17, 19, 24, 27–30) and colon cancer (25, 31, 32). Antibiotic treatment has been shown to inhibit bacterial populations responsible for deoxycholic acid formation and significantly decrease the cholesterol saturation index of bile (7). Despite the potential benefits of such treatment for individuals prone to cholesterol gallstone formation, the selection of antibiotic-resistant bacterial strains during long-term antibiotic administration precludes its effective use. The development of inhibitors specific for the bile acid 7α-dehydroxylation might be beneficial for preventing cholesterol gallstone formation. However, little is known about the genetics of bile acid 7α-dehydroxylation in intestinal bacteria.
Specific members of the genera Eubacterium and Clostridium are the only intestinal bacteria that have been shown to be capable of cholic acid 7α-dehydroxylation (12). Studies of Eubacterium sp. strain VPI 12708, an organism that can rapidly produce deoxycholic acid, identified a multistep pathway responsible for cholic acid 7α-dehydroxylation (8). Genetic analysis identified a bile acid-inducible operon (bai) that encodes enzymes required in this pathway. However, studies have shown that most cholic acid-7α-dehydroxylating intestinal bacteria belong to the genus Clostridium (33). More importantly, recent work found that Eubacterium sp. strain VPI 12708 bai genes cross-hybridized with DNA from other Eubacterium strains, but not with Clostridium strains tested (11). Based on this observation, Clostridium strains may have genetically distinct bai genes.
The present study was designed to identify the bile acid transporter gene from Clostridium sp. strain TO-931, a human fecal isolate. In addition, surrounding genes were cloned and sequenced to gain a better understanding of the genetics and enzymology of bile acid 7α-dehydroxylation in a Clostridium strain. Previous work showed that Clostridium sp. strain TO-931 had the highest cholic acid-7α-dehydroxylating activity of any intestinal bacteria tested, including Eubacterium sp. strain VPI 12708 (11). Knowledge of the bile acid 7α-dehydroxylation genetics in Clostridium species will allow for a better comparison of genes and gene products, which is necessary for developing specific bile acid 7α-dehydroxylation inhibitors.
MATERIALS AND METHODS
Isolation of chromosomal DNA.
Clostridium sp. strain TO-931 was kindly provided by Fusae Takamine (University of Ryukyus, Okinawa, Japan) and had been isolated from a human fecal sample. Cultures (50 ml) were grown in 100-ml volumes of peptone-yeast extract (PY) medium (18) supplemented with sucrose (4 g/liter), using anaerobically sealed serum bottles. Cells were collected by centrifugation (10,000 × g, 10 min) and suspended in 2-ml volumes of 0.9% saline. Cell suspensions were treated with 2 volumes of buffered phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol; Boehringer-Mannheim) and centrifuged (5,000 × g, 10 min). Phenol residue was removed by two equal-volume chloroform-isoamyl alcohol (24:1, vol/vol) extractions. Chromosomal DNA was precipitated with 1/20 volume of sodium acetate (3 M, pH 5.5) and 2.5 volumes of ice-cold ethanol and centrifuged. The DNA pellet was washed twice with ice-cold 70% ethanol, dried, dissolved in 250 μl of H2O, and stored at 2 to 4°C.
Oligonucleotide probe design.
Regions of the Eubacterium sp. strain VPI 12708 bile acid transporter sequence with homology to other transporters were scanned, and the nucleotide sequences having the least redundancy (<500) in the DNA sequence were used to design mixed oligonucleotide probes (50KM1 [5′-GARTAYCCNCARGARGAR-3′] and 50KM2 [5′-RCANACCCACATCCATNAC-3′]). Subsequent sequence-specific probes needed for cloning, sequencing, and PCR were identified by using Lasergene PrimerSelect software (DNASTAR Inc., Madison, Wis.). All oligonucleotides were commercially synthesized (Genosys Biotechnologies, The Woodlands, Tex.).
Detection of Clostridium sp. strain TO-931 bai genes.
Clostridium sp. strain TO-931 DNA (1 to 2 μg) was digested with AccI, AciI, BamHI, EcoRI, HinPI, NlaIII, Sau3AI, or XbaI (New England Biolabs, Beverly, Mass.). DNA fragments were separated by gel (1.0% agarose, Tris-acetate-EDTA buffer system) electrophoresis and transferred to a nitrocellulose membrane (Trans-Blot transfer medium; Bio-Rad Laboratories, Hercules, Calif.) for Southern hybridization analysis (13). DNA was cross-linked by using a UV Stratalinker 1800 (Stratagene, La Jolla, Calif.), and the nitrocellulose blots were hybridized for 12 h with probes labeled with [γ-32P]ATP (NEN, Boston, Mass.) by the use of T4 polynucleotide kinase. Blots were washed (13) and exposed to BioMax MS film (Kodak, Rochester, N.Y.).
Cloning of bai genes.
Chromosomal DNA was restriction enzyme digested and separated by agarose gel electrophoresis. DNA fragments were extracted from gel slices by using a Geneclean spin kit (Bio101, Vista, Calif.) and ligated into restriction enzyme-digested pUC19 (New England Biolabs), using T4 DNA ligase (New England Biolabs). Library Efficiency (Escherichia coli) DH5α competent cells (Gibco BRL, Gaithersburg, Md.) were used for DNA transformations. Clones were identified by colony hybridization analysis (3) by using probes that were labeled with [γ-32P]ATP by the use of T4 polynucleotide kinase (New England Biolabs). Clone identities were verified by restriction enzyme digestion and Southern hybridization analysis (13).
Difficult regions were cloned by the PCR technique, using a sequence-specific primer and a random primer (5′-GTTGGTGGCT-3′) to anchor downstream. The reaction mixture and conditions used were described previously (3) with the exception of the annealing temperature (35°C). The PCR products were separated by agarose gel electrophoresis, and their identities were verified by Southern hybridization analysis. The PCR products were cloned into the TA cloning vector (Stratagene).
DNA sequencing and sequence analysis.
Plasmid DNA was isolated from positive clones and sequenced by using a Dye-Terminator DNA sequencing kit (ABI Prism; Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif.). Sequence reactions were analyzed at the Medical College of Virginia-Virginia Commonwealth University Core Lab, using ABI Prism 373/375 sequence analyzers (PE Applied Biosystems). DNA sequences were submitted via the World-Wide Web to the National Institutes of Health for BLASTX analysis (1, 2). Cloned Clostridium sp. strain TO-931 bai gene sequences were arranged and managed by using Lasergene software (DNASTAR). Polypeptide analysis of BaiG was performed with the Lasergene software, and a transmembrane model was prepared by using the TMpred transmembrane prediction program operated via the World-Wide Web (http://ulrec3.unil.ch/software/TMRED_form.html).
RNA analysis and manipulations.
Clostridium sp. strain TO-931 was grown in PY broth with or without cholic acid (100 μM). RNA was isolated by using an RNeasy Midi kit (Qiagen, Chatsworth, Calif.), separated by 1% agarose gel electrophoresis, and transferred to nitrocellulose membranes (Bio-Rad Laboratories) for Northern hybridization analysis (3). The size of the mRNA transcript was determined by comparison to an RNA ladder (Ambion, Austin, Tex.).
RNA (5 to 10 μg) was precipitated with 3 M sodium acetate (1/20 volume) and ice-cold ethanol (2.5 volumes) at −20°C. The RNA pellet was dried and resuspended in RNase-free H2O with 32P-labeled oligonucleotide primer (5′-CATTCATATCGGTATTTTGCCTCCCTC-3′). RNA-primer mixtures were heated to 70°C for 10 min and allowed to cool slowly to room temperature to anneal the primers. Primers were extended by using SUPERSCRIPT II reverse transcriptase (Gibco BRL) at 42°C for 1 h. To determine the size of the extension product, DNA was manually sequenced using an fmol DNA PCR sequencing kit (Promega, Madison, Wis.) extended from the same 32P-labeled primer. The extension product and corresponding DNA sequencing products were separated by 6% acrylamide–40% urea gel electrophoresis (5). Following electrophoresis, the sequencing gel was dried and exposed to Kodak AR film. Primer extension and sequencing primers were 5′-end labeled with [γ-32P]ATP (NEN) as discussed above.
Nucleotide sequence accession number.
The nucleotide sequence of the Clostridium sp. strain TO-931 bai operon has been submitted to the GenBank database (accession no. ClosBai AF210152).
RESULTS
Clostridium sp. strain TO-931 bai gene identification.
The bai gene of Eubacterium sp. strain VPI 12708 encodes a bile acid transporter, and this gene exhibits homology to a large class of ATP-binding cassette transport proteins (20). Two redundant oligonucleotide primers (50KM1 and 50KM2) were based on potential membrane-spanning regions of the Eubacterium sp. strain VPI 12708 baiG gene product (20), and both sets hybridized to a single 1.0-kb DNA band in EcoRI-digested DNA from Clostridium sp. strain TO-931. Of 120 colonies, a single EcoRI clone was isolated using the 50KM1 probe set. The positive clone was sequenced, and the entire DNA sequence had 64.8% identity to the 5′ nucleotide sequence of the Eubacterium sp. strain VPI 12708 baiG gene.
Clostridium sp. strain TO-931 baiG gene analysis.
An open reading frame (ORF) similar in size and having 65% DNA sequence identity to the Eubacterium sp. strain VPI 12708 baiG gene was identified from an overlapping sequence generated from a PCR fragment and restriction enzyme (EcoRI and NlaIII)-generated clones of Clostridium sp. strain TO-931 DNA. The full-length polypeptide putatively encoded by Clostridium sp. strain TO-931 ORF had 71% identity and 81% similarity to the Eubacterium sp. strain VPI 12708 bile acid transporter. The polypeptide sequence of the Clostridium sp. strain TO-931 ORF had a hydrophobicity plot similar to that of the bile acid transporter (data not shown), and a two-dimensional model with 14 transmembrane segments was nearly identical to the bile acid transporter model proposed previously (20). Most variation between the peptide sequences was found to be in the C-terminal portion, specifically in the 6th external membrane loop between the 13th and 14th membrane-spanning helices.
Clostridium sp. strain TO-931 bai operon cloning and sequence analysis.
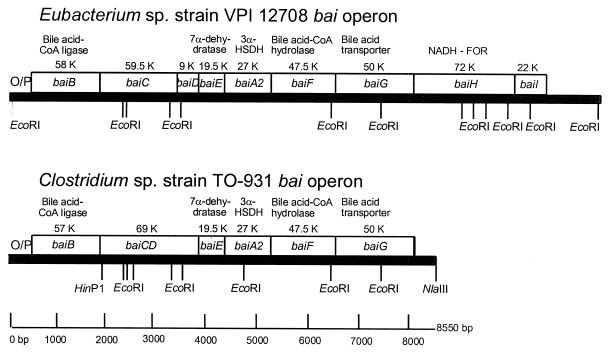
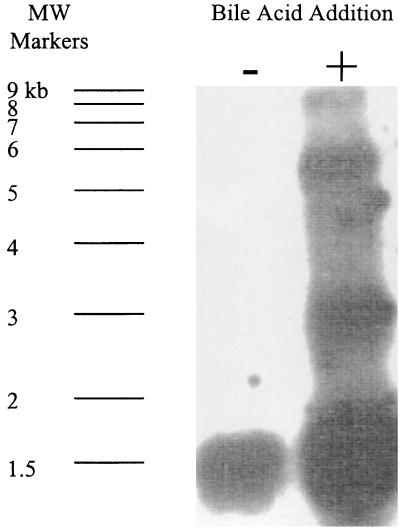
Nearly 9.2 kb of overlapping DNA sequence surrounding the baiG gene was combined from eight clones containing Clostridium sp. strain TO-931 DNA (Fig. 1). This large DNA sequence, which contained six ORFs and was expressed as a single mRNA of approximately 8 to 9 kb, was induced within 30 min following addition of 100 μM cholic acid to the growth medium (Fig. 2). This Clostridium sp. strain TO-931 bai operon had significant identity (Table 1) to the bai operon of Eubacterium sp. strain VPI 12708, and their gene orders were very similar (Fig. 1). No Eubacterium sp. strain VPI 12708 baiH-like or baiI-like genes were identified within 500 bp downstream of the baiG gene in the Clostridium sp. strain TO-931 operon. Interestingly, the baiC and baiD genes appeared to be encoded by a single continuous ORF rather than by two separate genes as observed for the Eubacterium sp. strain VPI 12708 bai operon (23). The intergene DNA sequences of Clostridium sp. strain TO-931 had little similarity to those observed in the Eubacterium sp. strain VPI 12708 bai operon and tended to be larger.
FIG. 1.
Overview of gene identity and organization for bile acid-inducible (bai) operons of Eubacterium sp. strain VPI 12078 and Clostridium sp. strain TO-931. No baiH or baiI gene was found downstream of Clostridium baiG. O/P, operator/promoter; FOR, flavin oxidoreductase; HSDH, hydroxysteroid dehydrogenase.
FIG. 2.
Northern blot analysis of the bai operon of Clostridium TO-931. Total RNA was isolated at time zero and at 30 min following addition (+) of cholic acid (50 μM) to the culture medium. In a control culture, bile acid was not added (−). Approximately 10 μg of RNA was loaded onto each lane and probed with the baiCD gene. MW Markers, molecular size marker positions.
TABLE 1.
Comparison of Eubacterium sp. strain VPI 12708 and Clostridium sp. strain TO-931 bai operon DNA sequences
| Gene or region | Length (bp), Eubacterium vs. Clostridium | Identity (%) | Protein | Enzymatic function | Identity (%) | Similarity (%) |
|---|---|---|---|---|---|---|
| Operator promoter | 218 vs 218 | 58 | ||||
| baiB | 1,563 vs 1,551 | 60 | BaiB | Bile acid-CoA ligase | 60 | 77 |
| baiB→baiCD (intergene) | 45 vs 95 | <10 | ||||
| baiCD | 1,920 vs 1,920 | 71 | BaiCD | Unknown | 83 | 91 |
| baiCD→baiE (intergene) | 43 vs 62 | <10 | ||||
| baiE | 501 vs 504 | 74 | BaiE | Bile acid 7α-dehydratase | 87 | 93 |
| baiE→baiA (intergene) | 33 vs 120 | <10 | ||||
| baiA | 747 vs 750 | 69 | BaiA | 3α-Hydroxysteroid dehydrogenase | 80 | 89 |
| baiA→baiF (intergene) | 63 vs 85 | <10 | ||||
| baiF | 1,281 vs 1,281 | 75 | BaiF | Bile acid-CoA hydrolase | 88 | 94 |
| baiF→baiG (intergene) | 5 vs 133 | <5 | ||||
| baiG | 1,434 vs 1,422 | 65 | BaiG | Bile acid transporter | 71 | 81 |
Comparison of the putative peptide sequences from each bai operon revealed significant identity, with nearly 90% similarity for most of the putative gene products (Table 1). The Clostridium sp. strain TO-931 putative gene product BaiCD aligned with both Eubacterium sp. strain VPI 12708 BaiC and BaiD putative gene products and had significant homology to Eubacterium sp. strain VPI 12708 BaiH (Fig. 3), a protein associated with an NADH:flavin oxidoreductase in Eubacterium sp. strain VPI 12708 (12).
FIG. 3.
Clustal alignment of peptide sequences for Eubacterium (Eub) sp. strain VPI 12708 bile acid-inducible proteins BaiC, BaiD, and BaiH and Clostridium (Clos.) sp. strain TO-931 bile acid-inducible protein BaiCD. Darkened residues (white letters on black background) denote identity and shaded residues (black letters on gray background) denote similarity between aligned sequences. The putative BaiCD peptide exhibits 46% identity and 51% similarity to BaiH, an enzyme with NADH:flavin oxidoreductase activity. Neither BaiC nor BaiD has been associated with an enzyme function.
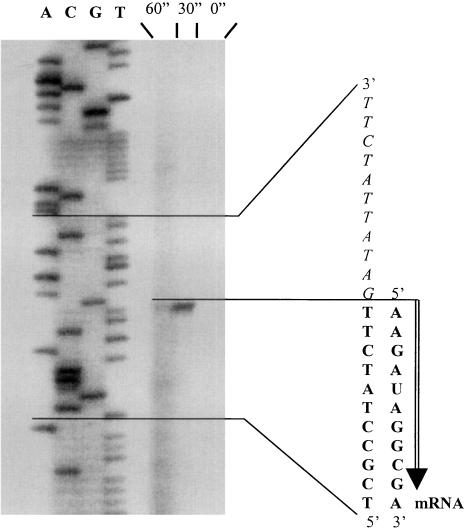
Promoter analysis.
The initial nucleotide for mRNA transcription was determined by primer extension analysis to lie 106 bases upstream from the Clostridium sp. strain TO-931 baiB gene (Fig. 4). Only bile acid-induced cultures yielded a primer extension product. The transcription initiation site in the DNA had an 8-bp sequence identical to that observed in Eubacterium sp. strain VPI 12708, and upstream were two regions identical to those observed in the Eubacterium sp. strain VPI 12708 promoter region (Fig. 5). In addition, several regions upstream from the putative promoter region are highly conserved and may be specific to bile acid regulation (5′-TTTGTCxxxxxATxxATTAGxTxTTxxxxxxxAAAAGGTx ATCTxTAxTxTTGTAAGAxxxCxxGxxATTAxCx-3′). The transcription initiation site for the Clostridium sp. strain TO-931 bai operon was surrounded by an inverted-repeat sequence (5′-TATC/AAGATA-3′) (Fig. 5) that was not observed in the Eubacterium sp. strain VPI 12708 bai operon DNA sequence (23).
FIG. 4.
Autoradiograph of primer extension analysis products. Lanes A, C, G, and T represent dideoxy nucleotide termination of Clostridium sp. strain TO-931 DNA sequence reactions. Lanes 60", 30", and 0" denote primer extension of Clostridium sp. strain TO-931 mRNA isolated from cultures at 60, 30, and 0 min, respectively, after induction with 100 μM cholic acid.
FIG. 5.
Alignment of bai operon promoter regions from Clostridium sp. strain TO-931 and Eubacterium sp. strain VPI 12708. Arrows denote transcription start sites as determined by primer extension analysis. The conserved regions are shaded with gray. The underlined sequences are the putative promoter binding (−10) sites. Clostridium sp. strain TO-931 DNA also had an inverted repeat (GATA/A/TATC) between the −10 site and the mRNA initiation site.
DISCUSSION
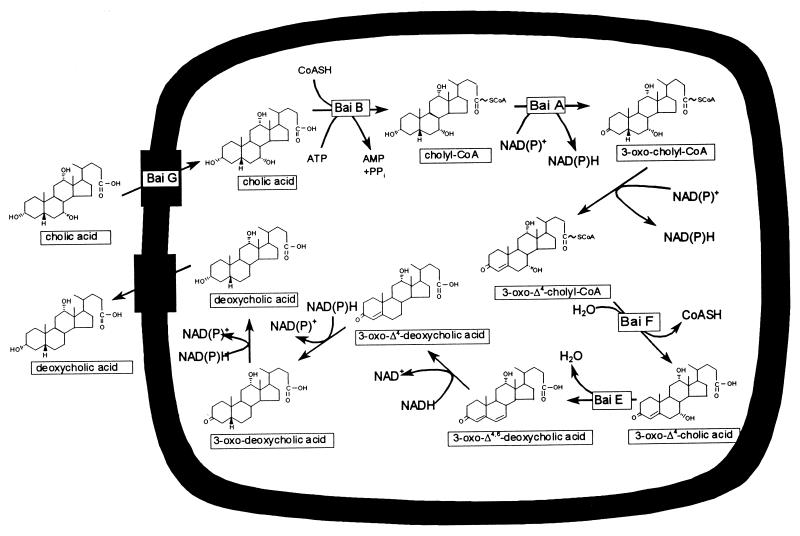
Primary bile acids are rapidly metabolized in the human colon via a 7α-dehydroxylation pathway (Fig. 6) that appears to be limited to certain strains of the genera Eubacterium and Clostridium (11). Bile acid 7α-dehydroxylation requires uptake of bile acids (20) and their conjugation to (21) coenzyme A (CoA) followed by two successive oxidation steps yielding a 3-oxo-Δ4-bile acid-CoA intermediate (4, 22). The intermediate appears to be deconjugated (34) and rapidly converted to a 3-oxo-Δ4,6-bile acid intermediate by 7α-dehydration (10). The 3-oxo-Δ4,6-bile acid intermediate is sequentially reduced to deoxycholic acid (11), and this end product is released from the cell.
FIG. 6.
Cholic acid 7α-dehydroxylation pathway in intestinal anaerobic bacteria. Bile acid-inducible (bai) gene products that participate in this pathway are indicated (also see Fig. 1). CoASH, coenzyme A-SH.
Many of the genes required for bile acid 7α-dehydroxylation have been identified in Eubacterium sp. strain VPI 12708 as part of a large (12-kb) bile acid-inducible operon (Fig. 1) (4, 23). These bile acid 7α-dehydroxylation genes from Eubacterium sp. strain VPI 12708 hybridized to DNA from other Eubacterium strains exhibiting bile acid 7α-dehydroxylation activity but failed to hybridize to a number of Clostridium strains (11). In addition, antibodies raised against purified bile acid 7α-dehydroxylation pathway enzymes from Eubacterium sp. strain VPI 12708 did not cross-react with proteins from bile acid-induced Clostridium strains (unpublished data). Although all intestinal bile acid 7α-dehydroxylation appears to proceed via a 3-oxo-Δ4-bile acid intermediate (8), these preliminary data suggested that the genes required for bile acid 7α-dehydroxylation in Eubacterium and Clostridium strains might be different.
Using a redundant oligonucleotide primer mix based on a membrane-spanning region of the Eubacterium sp. strain VPI 12708 bile acid transporter, we identified a baiG-like gene fragment in Clostridium sp. strain TO-931. Subsequent analysis identified a 1.42-kb ORF similar in size and sequence to the Eubacterium sp. strain VPI 12708 baiG gene. The Clostridium sp. strain TO-931 putative baiG gene product was found to have significant identity and similarity to the Eubacterium sp. strain VPI 12708 bile acid transporter (Table 1).
In Clostridium sp. strain TO-931, five putative ORFs were identified upstream of the baiG gene (Fig. 1), and each of these ORFs was found to exhibit significant identity to a Eubacterium sp. strain VPI 12708 bile acid 7α-dehydroxylation gene upstream of the baiG gene (Table 1). Clostridium sp. strain TO-931 bile acid 7α-dehydroxylation genes appears to be more AT biased (36% GC [versus 49% GC for the Eubacterium strain]) but were found to be organized in a similar fashion and to be similar in size to those identified in Eubacterium sp. strain VPI 12708 (Fig. 1). Nucleotide sequences complementary to Eubacterium sp. strain VPI 12708 baiB, baiC, baiD, baiE, baiA2, baiF, and baiG were observed, but no baiH or baiI genes were found. The baiH gene has been shown to encode an NADH:flavin oxidoreductase in Eubacterium sp. strain VPI 12708, but its function in bile acid 7α-dehydroxylation is unclear (12). No function has been assigned to the baiI gene product.
Although the two bacterial operons were found to have a high degree of individual gene identity, there are some differences. The baiC and baiD genes from Eubacterium sp. strain VPI 12708 were determined to be on overlapping but separate open reading frames (23). In Clostridium sp. strain TO-931, the baiC and baiD genes are fused into one continuous open reading frame that encodes a protein with 84% upstream and 75% downstream identity to the Eubacterium sp. strain VPI 12708 baiC and baiD gene products, respectively. The Clostridium sp. strain TO-931 baiCD gene aligned in its entirety with the Eubacterium sp. strain VPI 12708 baiH gene, and the putative gene products were shown to have 46% identity and 51% similarity (Fig. 4). Further analysis of the baiC, baiD, and baiE gene complements from both Clostridium sp. strain TO-931 and Eubacterium sp. strain VPI 12708 and the baiH and baiI genes from Eubacterium sp. strain VPI 12708 revealed a high degree of DNA sequence homology. These results suggest that gene duplication may have occurred in the Eubacterium sp. VPI 12708 bai operon. Because no baiH gene was found in the Clostridium sp. strain TO-931 bai operon, this enzyme function or a similar function may be associated with the Clostridium sp. strain TO-931 baiCD gene product. Further studies will be necessary to test this hypothesis.
In spite of the significant DNA sequence identity between the two bai operons, the intergene DNA sequences were determined to have little homology and often were found to be much larger in the Clostridium sp. strain TO-931 bai operon. The lack of identity, differences in size of the noncoding DNA, and AT bias suggest that there has been some genetic divergence. Despite the intergene differences, the operator/promoter regions upstream of the mRNA initiation site for both bai operons exhibit significant identity (Fig. 5). This putative bai promoter was shown to have little similarity to a proposed gram-positive promoter motif, but the latter proposed sequence appears to be based on genes expressed during the late-exponential and stationary phases of cell growth (14, 16, 26, 35, 36). Our bai promoter region may serve to regulate genes expressed in the presence of bile acids during exponential cell growth and, as a consequence of function, may represent a different class of promoters dependent on alternative sigma factors and/or auxiliary regulatory proteins.
In summary, we have shown that the bai operons of Clostridium sp. strain TO-931 and Eubacterium sp. strain VPI 12708 exhibit nearly 75% DNA sequence identity. More importantly, the putative gene products are homologous, with nearly 90% similarity for many of the proteins. Although the gene sequences may have changed over time, the putative proteins are highly conserved and probably have similar tertiary structures. This latter observation may be important for the development of bile acid 7α-dehydroxylation-inhibitory drugs.
ACKNOWLEDGMENTS
This work was supported by NIH program project grant P01-DK38030 to P.B.H. J.E.W. was supported by National Research Service award F32-DK09750 from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D P. Basic alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Baron S F, Hylemon P B. Biotransformation of bile acids, cholesterol, and steroid hormones. In: Mackie R I, White B A, editors. Gastrointestinal microbiology. Vol. 1. New York, N.Y: Chapman and Hall; 1997. pp. 470–510. [Google Scholar]
- 5.Baron S F, Franklund C V, Hylemon P B. Cloning, sequencing, and expression of the gene coding for bile acid 7α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. J Bacteriol. 1991;173:4558–4569. doi: 10.1128/jb.173.15.4558-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bazzoli F, Mazzella G, Parini P, Villanova N, Simoni P, Rossi L, Ronchi M, Festi D, Aldini R, Roda A, Roda E. Bile acid metabolism and enterohepatic dynamics in non-obese normolipidemic patients with cholesterol gallstones. Gastroenterology. 1991;100:A309. [Google Scholar]
- 7.Berr F, Kullak-Ublick G-A, Paumgartner G, Münzing W, Hylemon P B. Increased bacterial 7α-dehydroxylation as a mechanism for increasing deoxycholic acid input and supersaturated bile in cholesterol gallstone patients. Gastroenterology. 1996;111:1611–1620. doi: 10.1016/s0016-5085(96)70024-0. [DOI] [PubMed] [Google Scholar]
- 8.Björkhem I, Einarsson K, Melone P, Hylemon P. Mechanism of intestinal formation of deoxycholic acid from cholic acid in humans: evidence for a 3-oxo-Δ4-steroid intermediate. J Lipid Res. 1989;30:1033–1039. [PubMed] [Google Scholar]
- 9.Carey M C, Small D M. The physical chemistry of cholesterol solubility in bile: relationship to gallstone formation and dissolution in man. J Clin Investig. 1978;61:988–1026. doi: 10.1172/JCI109025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson J A, Mallonee D H, Björkhem I, Hylemon P B. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708. J Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 11.Doerner K C, Takamine F, LaVoie C P, Mallonee D H, Hylemon P B. Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63:1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklund C V, Baron S F, Hylemon P B. Characterization of the baiH gene encoding a bile acid-inducible NADH:flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J Bacteriol. 1993;175:3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopal-Srivastava R, Mallonee D H, White W B, Hylemon P B. Multiple copies of a bile acid-inducible gene in Eubacterium sp. strain VPI 12708. J Bacteriol. 1990;172:4420–4426. doi: 10.1128/jb.172.8.4420-4426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves M C, Rabinowitz J C. In vivo and in vitro transcription of the Clostridium pasteurianum ferrodoxin gene; evidence for 'extended' promoter elements in gram-positive organisms. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 15.Gray C H, Nicholson D C, Quincey R V. Fate of bile in the bowel. In: Heidel W, Code C, editors. Handbook of physiology. V. Washington, D.C.: American Physiological Society; 1968. pp. 2483–2505. [Google Scholar]
- 16.Hammond G A, Lyerly D M, Johnson J L. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb Pathog. 1997;22:143–154. doi: 10.1006/mpat.1996.0100. [DOI] [PubMed] [Google Scholar]
- 17.Hegardt F G, Dam H. The solubility of cholesterol in aqueous solutions of bile salts and lecithin. Z Ernaehrswiss. 1971;10:223–233. doi: 10.1007/BF02020933. [DOI] [PubMed] [Google Scholar]
- 18.Holdeman L V, Moore W E C. Anaerobe laboratory manual. 2nd ed. Blacksburg: Anaerobe Laboratory, Virginia Polytechnic Institute; 1972. [Google Scholar]
- 19.Hussaini S H, Pereira S P, Murphy G M, Dowling R H. Deoxycholic acid influences cholesterol solubilization and microcrystal nucleation time in gallbladder bile. Hepatology. 1995;22:1735–1744. [PubMed] [Google Scholar]
- 20.Mallonee D H, Hylemon P B. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallonee D H, Adams J L, Hylemon P B. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J Bacteriol. 1992;174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallonee D H, Lijewski M A, Hylemon P B. Expression in Escherichia coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol. 1995;30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 23.Mallonee D H, White W B, Hylemon P B. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J Bacteriol. 1990;172:7011–7019. doi: 10.1128/jb.172.12.7011-7019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus S N, Heaton K W. Deoxycholic acid and the pathogenesis of gallstones. Gut. 1988;29:522–533. doi: 10.1136/gut.29.4.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mower H F, Ray R M, Shoff R, Stemmerman G N, Nomura A, Glober G A, Kamiyama S, Shimada A, Yamakawa H. Fecal bile acids in two Japanese populations with different colon cancer risks. Cancer Res. 1979;39:328–331. [PubMed] [Google Scholar]
- 26.Nair R V, Green E M, Watson D E, Bennett G N, Papoutsakis E T. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J Bacteriol. 1999;181:319–330. doi: 10.1128/jb.181.1.319-330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsell K, Angelin B, Liljequist L, Einarsson K. Biliary lipid output and bile acid kinetics in cholesterol gallstone disease. Evidence for an increased hepatic secretion of cholesterol in Swedish patients. Gastroenterology. 1985;89:287–293. doi: 10.1016/0016-5085(85)90328-2. [DOI] [PubMed] [Google Scholar]
- 28.Paumgartner G, Sauerbruch T. Gallstones: pathogenesis. Lancet. 1991;338:1117–1121. doi: 10.1016/0140-6736(91)91972-w. [DOI] [PubMed] [Google Scholar]
- 29.Shaffer E A, Small D M. Biliary lipid secretion in cholesterol gallstone disease: the effects of cholecystectomy and obesity. J Clin Investig. 1977;59:828–840. doi: 10.1172/JCI108705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoda J, He B F, Tanaka N, Matsuzaki Y, Osuga T, Yamamori S, Miyazaki H, Sjovall J. Increase of deoxycholate in supersaturated bile of patients with cholesterol gallstone disease and its correlation with de novo synthesis of cholesterol and bile acids in the liver, gallbladder emptying, and small intestine transit. Hepatology. 1995;21:1291–1302. [PubMed] [Google Scholar]
- 31.Vlahcevik Z R, Heuman D M, Hylemon P B. Physiology and pathophysiology of enterohepatic circulation of bile acids. In: Zakim D, Boyer T D, editors. Hepatology, a textbook of liver disease. W. B. Philadelphia, Pa: Saunders; 1996. pp. 376–401. [Google Scholar]
- 32.Weinstein I B. Nonmutagenic mechanisms in carcinogenesis: role of protein kinase C in signal transduction and growth control. Health Perspect. 1991;93:175–179. doi: 10.1289/ehp.9193175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells J E, Berr F, Thomas L A, Dowling R A, Hylemon P B. Isolation and characterization of cholic acid 7α-dehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol. 2000;32:4–10. doi: 10.1016/s0168-8278(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 34.Ye H Q, Mallonee D H, Wells J E, Björkhem I, Hylemon P B. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J Lipid Res. 1999;40:17–23. [PubMed] [Google Scholar]
- 35.Young M, Minton N P, Staudenbauer W L. Recent advances in the genetics of clostridia. FEMS Microbiol Rev. 1989;63:301–326. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Melville S B. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J Bacteriol. 1998;180:136–142. doi: 10.1128/jb.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]