Abstract
Genes encoding l-arginine biosynthetic and transport proteins have been shown in a number of pathogenic organisms to be important for metabolism within the host. In this study we describe the cloning of a gene (Rv0522) encoding an amino acid transporter from Mycobacterium bovis BCG and the effects of its deletion on l-arginine transport and metabolism. The Rv0522 gene of BCG was cloned from a cosmid library by using primers homologous to the rocE gene of Bacillus subtilis, a putative arginine transporter. A deletion mutant strain was constructed by homologous recombination with the Rv0522 gene interrupted by a selectable marker. The mutant strain was complemented with the wild-type gene in single copy. Transport analysis of these strains was conducted using 14C-labeled substrates. Greatly reduced uptake of l-arginine and γ-aminobutyric acid (GABA) but not of lysine, ornithine, proline, or alanine was observed in the mutant strain compared to the wild type, grown in Middlebrook 7H9 medium. However, when the strains were starved for 24 h or incubated in a minimal salts medium containing 20 mM arginine (in which even the parent strain does not grow), l-[14C]arginine uptake by the mutant but not the wild-type strain increased strongly. Exogenous l-arginine but not GABA, lysine, ornithine, or alanine was shown to be toxic at concentrations of 20 mM and above to wild-type cells growing in optimal carbon and nitrogen sources such as glycerol and ammonium. l-Arginine supplied in the form of dipeptides showed no toxicity at concentrations as high as 30 mM. Finally, the permease mutant strain showed no defect in survival in unactivated cultured murine macrophages compared with wild-type BCG.
The mycobacteria are distinguishable from most other organisms on the basis of their low permeability. Cell wall structural analysis (41) and permeability studies (18, 28) have provided an increased understanding of this characteristic. However, few of the mycobacterial transporters mediating the uptake of nutrients have been characterized at the genetic, molecular, or biochemical levels. There are few recent studies of nutrient transport by mycobacteria (9; for a review, see reference 18), and, in particular, there are no studies of the role of nutrient transport in the intracellular survival of mycobacteria. The nature and availability of carbon, nitrogen, and energy sources within the macrophage can best be studied with genetic mutants altered in intermediary metabolism and transport. This study presents a genetic approach to the transport of l-arginine in mycobacteria.
Many microorganisms use l-arginine as a source of energy and/or nitrogen, and the pathways of l-arginine biosynthesis and utilization are well understood in some cases. At least five pathways of l-arginine catabolism have been identified in microorganisms, listed here by the first enzyme to act upon the substrate: arginase, arginine deiminase, arginine decarboxylase, arginine succinyl transferase, and arginine oxidase. l-Arginine metabolism has not previously been examined in the slow-growing mycobacteria, but scrutiny of the genomic sequence of Mycobacterium tuberculosis (16) reveals the presence of two of these five enzymes, arginine deiminase (Rv1001) and arginine decarboxylase (Rv2531c). Arginine decarboxylase has been studied in the context of regulation of pyrimidine synthesis in M. smegmatis (2, 3, 5, 40).
In the arginase pathway, l-arginine is converted to urea and ornithine. Urea is subsequently converted to NH3 and CO2 by urease. There is no apparent homolog of arginase in the Sanger database, but homologs of genes of subsequent enzymes in the arginase pathway are present in the H37Rv genome. In addition, the urease genes of BCG and M. tuberculosis have been cloned and characterized (15, 46). By constructing a strain lacking the urease gene, it was demonstrated that ureolytic activity was not essential to BCG Δure grown in vitro. However, a slight decrease in the multiplication and persistence of the mutated strain compared with wild-type BCG was observed in the lungs of infected mice (47).
l-Arginine transport is an important aspect of arginine metabolism and is regulated in concert with l-arginine catabolic enzymes in many bacterial systems. Several different classes of permeases are responsible for l-arginine transport among the bacteria: the existence within a single organism of multiple transport systems for this amino acid attest to its importance. In Escherichia coli and Salmonella, the major l-arginine permease is a member of the binding protein-dependent family of transporter systems, with three separate periplasmic binding proteins of differing specificities (24). The first binds l-lysine, l-arginine, and l-ornithine (11, 48); the second binds l-arginine and l-ornithine (13); and the third binds only l-arginine (48, 53).
In Bacillus subtilis, the rocE and rocC genes encode putative arginine permeases and are homologous to each other (23, 42). The sequence of these permease genes probably classifies them as members of the amino acid-polyamine-organocation superfamily, single-protein membrane carriers characterized by 12 or 14 transmembrane α-helices (37). To study l-arginine transport in mycobacteria, the M. tuberculosis sequence was searched for predicted open reading frames (ORFs) with homology to the B. subtilis l-arginine transport proteins. Two homologs were identified, Rv0522 and Rv2320c.
Here, Rv0522 was cloned from BCG and used to construct a strain lacking the permease encoded by this ORF. The deletion strain and its wild-type parent are characterized with respect to transport, growth properties, and survival in a murine macrophage cell line.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were grown in Luria-Bertani medium, and mycobacterial strains were grown in Middlebrook medium [per liter, (NH4)SO4, 0.5 g; l-glutamic acid, 0.5 g; sodium citrate, 0.1 g; pyridoxine, 0.001 g; biotin, 0.0005 g; Na2HPO4, 2.5 g; KH2PO4, 1.0 g; ferric ammonium citrate, 0.04 g; MgSO4, 0.05 g; CaCl2, 0.0005; ZnSO4, 0.001; CuSO4, 0.001 g]. Middlebrook 7H9 (liquid) and 7H11 (1.5% agar) media (Difco) were supplemented with glycerol (0.5% vol/vol) and ADC supplement (0.5% bovine serum albumin, fraction V [Boehringer Mannheim], 0.2% dextrose, 0.85% NaCl). Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml, kanamycin and streptomycin, 50 μg/ml for E. coli and 20 μg/ml for BCG; and hygromycin, 150 μg/ml for E. coli and 50 μg/ml for BCG. The minimal medium used was composed of basal salts (0.1% KH2PO4, 0.25% NaH2PO4, 0.5% NH4Cl, 0.2% K2SO4) medium supplemented with glycerol (0.5%). Sauton's medium was used without amino acid supplements (17). Where applicable, the nitrogen (NH4Cl) and/or carbon (glycerol and/or dextrose) sources were omitted. All liquid cultures of BCG were supplemented with 0.05% Tween 80 (Sigma Chemicals).
TABLE 1.
Strains and plasmids used in this study
| Designation | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 26 |
| GM48 | dam-3 dcm-6 thr-1 ara-14 tonA31 lacY1 tsx-78 glnV44 galK2 galT22 thi-1 | 38 |
| M. bovis BCG strains | ||
| BCG | Pasteur strain | Difco |
| AS1 | BCG (Rv0522 interrupted with Kan-Str) | This study |
| AS2 | AS1 complemented with a wild-type copy of Rv0522 (pAS7) | This study |
| Plasmids | ||
| pGEM3Zf− | Ampr; multiple-cloning-site vector | |
| pMV305 | Kanr; E. coli-mycobacterium integrating shuttle vector | 32 |
| PSM240 | Kan-Str cassette in pBEN | I. Smith |
| pMV261 | Kanr; E. coli-mycobacterium shuttle expression vector with hsp60 promoter | 51 |
| pJB3 | pUC19 containing Hygr from pIJ963 as a SpeI fragment | J. Berger |
| pAS5 | pGEM3Zf− with a 6-kb EcoRI fragment from cosmid 121 of the pYUB18 BCG cosmid library | This study |
| pAS6 | pAS5 with a 3.5-kb BamHI Kan-Str cassette from pSM240 in the BclI site of pAS5 | This study |
| pAS7 | pMV305 with the 6-kb EcoRI fragment from pAS5 and a HygrSpeI fragment from pJB3 in the NheI site | This study |
| pYUB18 | Kanr; cosmid library vector | 27 |
Cloning and DNA manipulations.
Plasmid DNA preparations, restriction endonuclease digestions, alkaline phosphatase treatments, ligations, transformations, and other DNA manipulations were performed by standard procedures (36). For electroporation of BCG, the cells were grown until the culture reached a turbidity (optical density at 600 nm [OD600]) of about 0.6 and then harvested at 4,000 × g for 10 min. The pellet was washed with 10% glycerol and centrifuged again at 4,000 × g for 10 min. The procedure was repeated two more times before resuspension of the pellet in 1/10 volume of 10% glycerol. All manipulations were carried out at 37°C. DNA (3 to 5 μg) was added to 0.5 ml of cells in a 0.4-cm electroporation cuvette (BTX). The cuvette was subjected to a single pulse using the Bio-Rad Gene Pulser set at 2.5 kV and 25 μF, with the pulse controller resistance set at 1,000. The contents of the cuvette were diluted into 5 ml of 7H9 medium and incubated overnight at 37°C. After incubation, the entire 5 ml was centrifuged and plated onto 7H10-antibiotic plates. Transformants appeared after incubation at 37°C for 3 to 4 weeks.
Construction of BCG Rv052 deletion strain.
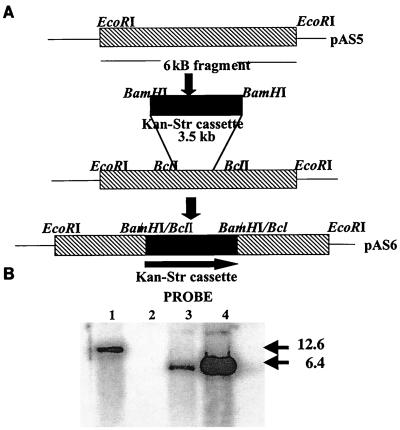
PCR analysis to screen the BCG genomic library to isolate a cosmid containing the B. subtilis rocE homolog was performed using the PCR kit from Boehringer Mannheim Biochemicals. The primers used were 5′ ATCGTGATCTTCTTCGTCGG 3′ and 5′ ATGATCACGCACAGGAATCC 3′. The cosmid DNA was digested with a number of restrictions enzymes, and a Southern analysis using the PCR product as a probe revealed the presence of a 6-kb EcoRI fragment bearing the homolog. This fragment was then subcloned into pGEM3Zf− and is called pAS5 (see Fig. 1). This was then transformed into a dam E. coli strain, GM48. Transformation into GM48 permitted digestion with the enzyme BclI, which released a 191-bp fragment. A Kan-Str cassette as a BamHI fragment from plasmid pSM240 was then inserted into the BclI site of pAS5 to create pAS6. This construct was then used for allelic exchange of the homolog, the Rv052 gene in BCG.
FIG. 1.
Construction of Rv052 deletion strain. (A) Cloning strategy for construction of AS1. See Materials and Methods for details. (B) Southern analysis of genomic DNA from BCG and AS1 strains. Genomic DNA from a single-crossover recombinant (lane 1), wild-type BCG (lane 2), and mutant AS1 (lane 3) and plasmid DNA from pAS6 (lane 4) were digested with EcoRI and probed with the Kan-Str casette.
PCR and Southern analysis were used to differentiate between single- and double-crossover events (see Fig. 1). BCG genomic DNA was isolated using N-acetyl-N,N,N-trimethylammonium bromide) (Sigma) as previously described (18). PCR was performed using the Expand Long Template Kit (Boehringer Mannheim) as specified by the manufacturer. The primers 5′ AGCCACATCCGTACCCCCAG 3′ and 5′ CACGCATCGTGATCTTCTTCG 3′ flank the Kan-Str cassette such that single crossovers would show fragments of 471 bp of the wild-type copy of the gene and the 3.5-kb fragment containing the Kan-Str marker. True allele replacements would lack the small band (deleted by recombination out of the genome) and would show only the 3.5-kb fragment. The PCR results were confirmed by Southern analysis of the double recombinant with the Kan-Str marker as a probe. Southern hybridization was performed by the method of Maniatis et al. (36). The Southern analysis also shows that the Kan-Str cassette inserted only once in the genome.
Uptake assays.
Cells were grown to mid-log phase, washed three times with basal salts medium plus Tween 80 at 0.05%, and concentrated in basal salts medium approximately fivefold to a final OD600 of 3.0. The cell suspensions (1 ml) were warmed to 37°C with shaking. The culture was treated with rifampin (200 μg/ml) for 10 min prior to initiation of an uptake assay to block transcription and subsequently protein synthesis; this prevented the unlimited incorporation of radiolabeled amino acids into protein. The uptake reaction was initiated by the addition of radiolabeled substrate plus unlabeled substrate at the specific activities (usually 40 to 50 μCi/μmol) and to the final concentrations (usually 100 μM) described in the legends of the figures. Incorporation was terminated by removal of 0.1-ml samples at the indicated time to filters (Whatman GF/F; 0.45 μm) prewetted with BS medium. The cells were rinsed quickly (within 10 to 15 s) three times with 7 ml of ice-cold basal salts medium plus Tween 80 on a Hoeffer 10-place manifold with air vacuum. Filters with cells thereon were transferred to vials containing 5 ml of scintillation fluid for determination of radioactivity. The counts per minute were normalized to milligrams of protein per 0.1-ml aliquot for each cell suspension; protein was determined by the Bio-Rad protein assay.
For analysis of uptake by cells resuspended in different media, care was taken to ensure that the cultures used for uptake studies were at similar densities before the washing and concentration steps. This was essential when cells were resuspended in a medium that does not support measurable growth (minimal salts plus amino acid or γ-aminobutyric acid [GABA]) and compared with cells resuspended in Middlebrook medium.
To measure the effect of exogenous l-arginine on [3H]uracil incorporation, 0.5 μCi of [3H]uracil (38.5 Ci/mmol) (New England Nuclear) and peptide were added simultaneously to the cells to initiate the experiment. After 16 h, the cultures were precipitated in 10% trichloroacetic acid (TCA) (Sigma) at 4°C for 20 min, filtered, and washed three times with 5 ml of cold 10% TCA over filters (Whatman GF/F; 0.45-μm pore size) prewetted with 10% TCA on a Hoeffer 10-place manifold with air vacuum (as above).
Macrophage survival assays.
J774.1 cells were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal calf serum (Sigma), amino acids (BioWhittaker), and l-glutamine (Sigma). The cells were subcultured into 96-well plates (1.5 × 105 cells per well) for 6 to 12 h. Then 2 × 106 CFU of logarithmically growing bacteria were washed in macrophage medium and placed in the wells. After 4 to 6 h of attachment, the wells were rinsed three times in macrophage medium. The numbers of intracellular bacteria was determined by removing the supernatant of three wells in parallel, lysing the macrophages in double-distilled H2O ddH2O, diluting, and plating for CFU on 7H11 plates with the appropriate antibiotics. The supernatants were checked for the presence of extracellular bacteria at every time point, and the number of extracellular bacteria remained in the range of 1 to 5% of that of intracellular organisms. Triplicate determinations were made for each time point, and the experiment was performed five times.
RESULTS
Cloning and interruption of the rocE permease homolog from BCG.
The rocE gene of B. subtilis encodes a major l-arginine permease. Its expression is induced by l-arginine in the medium, controlled at the level of transcription by the product of the rocR gene, a member of the NtrC/NifA family of regulators. Using the amino acid sequence of the B. subtilis rocE gene, the Sanger TB database was analyzed for ORFs with homology to the rocE gene. The strongest homology was exhibited by Rv0522 (31% identity and 53% similarity). The sequence of this predicted ORF in turn shows 44.3% identity to the GABA permease gene (gabP) of E. coli and 20% identity to gabP of B. subtilis. In B. subtilis, the rocE gene is part of the rocDEF operon bearing the genes for l-arginine catabolism. In contrast, Rv0522 is not part of an operon and is flanked by divergent ORFs of unknown function.
Primers derived from the Sanger database (16) were used to screen a genomic library of BCG (Pasteur) DNA in pYUB18 (27) for the presence of Rv0522 sequences. A cosmid carrying the BCG homolog of Rv0522 was identified (Fig. 1). From this cosmid, an EcoRI fragment of 6 kb bearing the BCG Rv0522 homolog was cloned into pGEM3Zf to create pAS5 (Table 1). A 191-bp BclI fragment in the ORF was removed and replaced with an antibiotic cassette containing kanamycin and streptomycin resistance markers to create pAS6. This final construct was linearized with BamHI and electroporated into BCG. Transformants resistant to both kanamycin and streptomycin were selected. The candidates for allele replacements were first screened by PCR, using oligonucleotide primers flanking the marker insertion site (see Materials and Methods). Southern blot analysis confirmed the structure of genomic DNA representing the wild-type strain and the allele replacement (double crossover) strain, AS1 (Fig. 1).
To complement the deletion, a wild-type copy of the Rv0522 gene was inserted in single copy into AS1. The mycobacteriophage L5 attachment site (attB) was used for integration of pAS7, the pMV305 plasmid carrying the attP site of L5 (32) and a wild-type copy of the BCG Rv0522 gene. The complemented mutant strain was named AS2.
Uptake properties of AS1 (ΔRv0522).
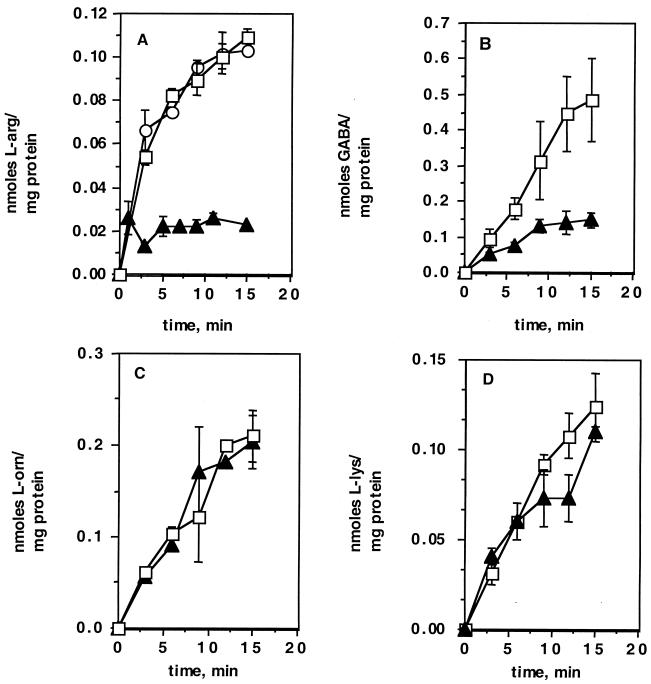
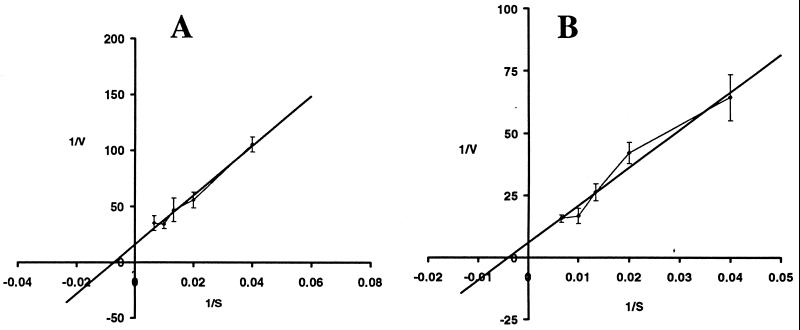
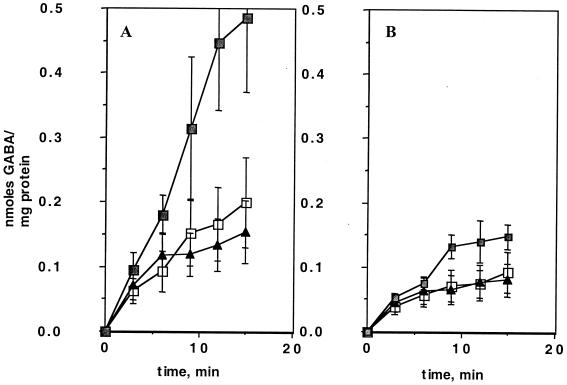
AS1 and its wild-type parent were analyzed for uptake of various l-amino acids by using radiolabelled substrates. The cells were grown in Middlebrook medium (7H9 plus glycerol, ADC supplement, and 0.05% Tween). The mutant strain showed a significant decrease in uptake of l-[14C]arginine (90% reduction) (Fig. 2A). In addition, the mutant showed a clear defect (75% reduction) in uptake of [14C]GABA compared with wild-type cells (Fig. 2B). Neither l-[14C]arginine nor [14C]GABA uptake was completely abolished. This was not unexpected, as most bacteria have multiple transporters for l-arginine. There was no difference between the wild type and mutant in uptake of the structurally related amino acids l-[14C]ornithine (Fig. 2C) or l-[14C]lysine (Fig. 2D). The complemented mutant strain, AS2, contained a wild-type copy of the Rv0522 gene supplied in trans that fully complemented the l-[14C]arginine (Fig. 2A) and [14C]GABA (data not shown) uptake defects. Comparison of the uptake of structurally unrelated amino acids, l-alanine and l-proline, indicated that there were no differences between the wild-type and AS1 strains (data not shown). Finally, the apparent Kms for transport of l-[14C]arginine and [14C]GABA were calculated from Lineweaver-Burk analyses (Fig. 3) and found to be 250 and 165 μM, respectively.
FIG. 2.
Uptake of 14C-labeled amino acids by wild-type BCG (□), AS1 (▴), and AS2 (○). (A) Uptake of l-[14C]arginine. (B) Uptake of [14C]GABA. (C) Uptake of l-[14C]ornithine. (D) Uptake of l-[14C]lysine.
FIG. 3.
Lineweaver-Burk analysis of l-arginine and GABA uptake. Double inverse plots of [14C]GABA (A) and l-[14C]arginine (B) are shown. Uptake proceeded for 2 min at the concentrations shown on the x axis. The apparent Kms were calculated from the x axis intercept. 1/s = (nanomoles of substrate/milligram of protein/minute)−1; 1/v = (micromoles of substrate)−1.
l-[14C]arginine uptake by wild-type BCG and AS1.
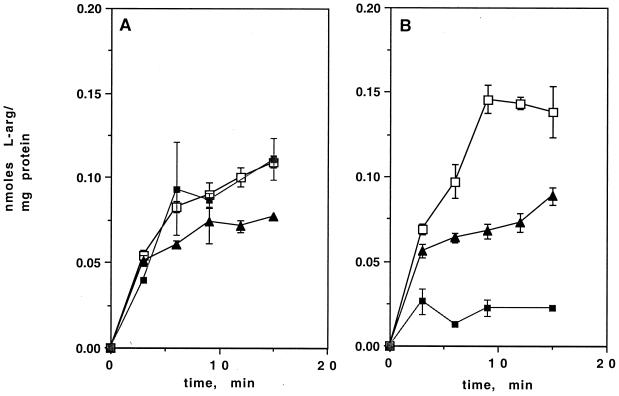
In other bacteria, expression of l-arginine catabolic and biosynthetic operons is regulated by the presence or absence of exogenous l-arginine (7, 13, 23, 53). This regulation is often mediated by l-arginine acting as a corepressor in concert with the ArgR/AhrC protein as a repressor (34). To examine the effect of exogenous l-arginine on uptake of l-[14C]arginine in wild-type BCG, cells in balanced growth in Middlebrook medium were washed and incubated in minimal salts medium (see Materials and Methods) with 20 mM l-arginine as the sole carbon and nitrogen source. After 24 h at 37°C, uptake of l-[14C]arginine by cells incubated in l-arginine alone was only slightly reduced (25%) in comparison to that by wild-type cells growing in Middlebrook medium (Fig. 4A). The effect of starvation for carbon and nitrogen on l-[14C]arginine uptake by wild-type BCG was also examined. After 2 h (data not shown) or 24 h (Fig. 4A) of starvation in basal salts lacking sources of carbon and nitrogen, l-[14C]arginine uptake by wild-type cells was unchanged.
FIG. 4.
Comparison of uptake of l-[14C]arginine by BCG and AS1 under different nutritional conditions. Uptake of l-[14C]arginine by cells growing in Middlebrook medium (■), after overnight starvation (no carbon and nitrogen source) (□), or after overnight treatment with 20 mM l-arginine as the sole carbon and nitrogen source (▴) is shown. (A) Uptake by wild-type BCG. (B) Uptake by AS1.
Mid-log-phase AS1 cells were washed and incubated for 2 h (data not shown) or 24 h in minimal salts with 20 mM arginine. Figure 4B shows that in contrast to the reduction seen in wild-type BCG, l-[14C]arginine uptake by the AS1 mutant was strongly increased over the levels found in AS1 grown in 7H9 medium. The results were the same after 2 h in 20 mM arginine. This suggests that exogenous l-arginine in the medium may induce a permease other than that encoded by Rv0522.
Uptake of l-[14C]arginine by starved AS1 cells was also measured (Fig. 4B). Surprisingly, under these conditions, the mutant strain showed levels of uptake higher than under any other conditions, including those exhibited by the wild type after growth on Middlebrook medium.
[14C]GABA uptake by wild-type BCG and AS1.
To examine the effect of exogenous GABA on [14C]GABA uptake in BCG, wild-type cells in balanced growth in Middlebrook medium and cells washed and incubated in minimal salts medium with 20 mM GABA as the sole carbon and nitrogen source were compared. Figure 5A shows that there was a 70% reduction in [14C]GABA uptake by wild-type cells after exposure to 20 mM GABA. The results were identical after only 2 h of incubation in 20 mM GABA (data not shown).
FIG. 5.
Comparison of uptake of [14C]GABA by BCG and AS1 under different nutritional conditions. Uptake of [14C]GABA by cells growing in Middlebrook medium (■), after overnight starvation (no carbon or nitrogen source) (□), or after overnight treatment with 20 mM GABA as the sole carbon and nitrogen source (⧫) is shown. (A) Uptake by wild-type BCG. (B) Uptake by AS1.
The relative contribution of the Rv0522 permease to this reduction in GABA uptake was evaluated by measuring the effect of GABA exposure on [14C]GABA uptake by AS1 cells. Figure 5B shows that uptake of [14C]GABA was reduced 60% in AS1 after 24 h of incubation in 20 mM GABA, in comparison with mutant cells in Middlebrook medium.
Cultures growing in Middlebrook medium were washed and incubated in minimal salts with no carbon or nitrogen source. The effects of this starvation on [14C]GABA uptake by wild-type BCG were evaluated and shown in Fig. 5A. There was a 70% reduction in uptake of [14C]GABA by wild-type cells after starvation, similar to that exhibited by wild-type cells after GABA exposure. AS1 cells also showed a reduction comparable to that exhibited after GABA exposure (Fig. 5B). Thus, unlike GABA uptake in B. subtilis (21), GABA uptake by wild-type BCG was not induced by starvation for carbon and nitrogen, and was, in fact, slightly repressed. Furthermore, the reduction shown in [14C]GABA uptake by AS1 suggested that a wild-type uptake activity not affected by the deletion of Rv0522 was responsive to carbon and nitrogen starvation.
l-Arginine utilization by BCG.
Growth of wild-type BCG and AS1 cells in minimal arginine medium was measured. The cells were first grown to the mid-logarithmic growth phase in standard Middlebrook medium. They were washed and resuspended at an OD600 of 0.2 in various minimal media (basal salts medium or Sauton's medium [see Materials and Methods]) containing at 1 or 20 mM l-arginine as the sole carbon and/or sole nitrogen source. Surprisingly, neither wild-type nor mutant cells were capable of growing in either medium with l-arginine as the sole carbon or nitrogen source. In basal salts medium or Sauton's medium supplemented with glycerol (0.5%) and ammonium chloride (0.5%), wild-type cells grew at rates comparable to those seen for Middlebrook 7H9-grown cells. The following amino acids were tested as sources of either carbon or nitrogen to support the growth of wild-type BCG: l-histidine, l-lysine, l-ornithine, GABA, l-alanine, and l-proline. In all cases, single amino acids were incapable of supporting growth. This is in marked contrast to the observation that a wide range of amino acids and di- and tripeptides at a concentration of 2 to 5 mM are capable of supporting the growth of wild-type M. smegmatis in minimal medium as either the sole carbon or nitrogen source (9, 45).
Effect of exogenous arginine on wild-type BCG.
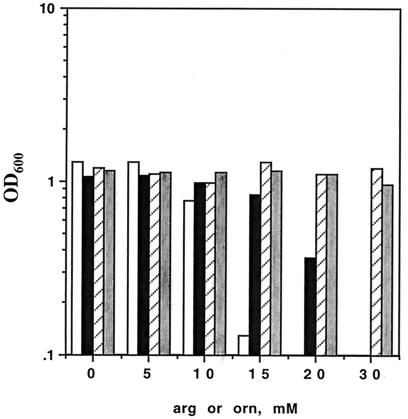
Wild-type BCG was grown in Middlebrook 7H9 medium, containing glycerol and ammonium, in the presence of exogenous l-arginine at concentrations ranging from 0 to 40 mM (Fig. 6). Note that in this experiment, the primary sources of carbon and nitrogen (glycerol and ammonium, respectively) provided by Middlebrook medium are optimal for growth of the slow-growing mycobacteria (45). Surprisingly, at 15 mM arginine, there was some inhibition of growth, and at 20 mM, growth was completely inhibited. Structurally related amino substrates, such as l-ornithine (Fig. 6), l-lysine (results not shown), and GABA (results not shown), and unrelated amino acids (l-proline and l-alanine [results not shown]) at the same concentrations showed no inhibition of growth; therefore, this growth inhibition was specific for arginine. Furthermore, growth inhibition by l-arginine was reduced in AS1 (Fig. 6), as would be predicted from the reduced arginine transport in AS1 cells (Fig. 3A).
FIG. 6.
Effect of exogenous l-arginine or l-ornithine on growth of wild-type and mutant BCG. Exponentially growing cells were washed and resuspended at an OD600 of 0.1 in Middlebrook 7H9 medium in the presence of exogenous l-arginine at the concentrations indicated. The OD600 was measured after 4 days of growth at 37°C. From left to right for each concentration, wild-type BCG plus l-arginine (open bars), AS1 plus l-arginine (solid bars), wild-type BCG plus l-ornithine (hatched bars), and AS1 plus l-ornithine (shaded bars). The experiment is representative of three similar experiments.
Amino acids can also be supplied to cells in the form of peptides. Indeed, Marquis et al. showed that in culture, threonine auxotrophs of Listeria monocytogenes grow poorly on free threonine and quite well on threonine-containing peptides. These same auxotrophs showed no difference in growth rate within threonine-starved J774 macrophages, suggesting that threonine-containing peptides are available for intracytoplasmic growth (39). In BCG, there is no toxicity of exogenous l-arginine when supplied at 20 or 30 mM in the form of l-arginyl-l-glutamate or l-arginyl-l-asparagine (data not shown).
High concentrations of exogenous l-arginine are cytocidal.
To determine whether the effect of exogenous arginine on the growth of wild-type BCG is cytostatic or cytocidal, the cultures described in Fig. 6 were plated to determine CFUs. There was no effect of 10 mM arginine on viability, but the culture containing 20 mM arginine contained no viable cells (data not shown). To confirm this observation, a labeling assay (14) was used to evaluate the metabolic activity of wild-type BCG exposed to exogenous arginine. [3H]uracil incorporation into precipitable macromolecules was inhibited by 50% in wild-type BCG growing in rich medium and incubated in 20 mM l-arginine for 24 h (data not shown). In contrast, the addition of 20 mM l-arginine to BCG resuspended in basal salts medium (starvation conditions) had little effect (10% inhibition) on [3H]uracil incorporation. Thus, the effect of exogenous l-arginine on [3H]uracil incorporation appears to depend on the nutritional state of the culture.
Survival of strain AS1 in cultured murine macrophages.
Roles for genes involved in l-arginine transport (30) and metabolism (35) have been implicated in macrophage infection studies (see Discussion). Wild-type BCG, AS1, and AS2 were evaluated for survival in unactivated J774.1 macrophages. No differences were found among the three strains.
DISCUSSION
Little is known about the regulation of amino acid transport and metabolism in mycobacteria. As a first step to understanding l-arginine metabolism in BCG, we screened the Sanger database for ORFs with homology to the arginine permeases of E. coli, Pseudomonas spp., and B. subtilis. The highest homology pointed to Rv0522, which was homologous to the rocE arginine permease of B. subtilis. Rv0522 also showed 20% identity and 19% similarity to the B. subtilis gabP gene. The BCG homolog of Rv0522 was cloned, interrupted, and crossed onto the chromosome of BCG by gene replacement. The mutant thus constructed, AS1, showed decreased uptake of both l-[14C]arginine and [14C]GABA.
Arginine metabolism has been widely studied in bacteria (see the introduction), and a range of regulatory systems controls the arginine catabolic genes, including those encoding transporters. In the enterics, l-arginine uptake systems are either repressed or unaffected by exogenous l-arginine: the ArgR protein functions largely in the repression of the arginine biosynthetic operons (34). In P. aeruginosa and B. subtilis, exogenous l-arginine induces l-arginine uptake (7, 43).
There are no published reports of l-arginine or GABA transport by the slow-growing mycobacteria. l-arginine transport by the fast-growing species M. phlei has been measured as part of a larger study of the energetics of amino acid transport (45, 54). From our data, we estimate initial uptake rates of l-arginine in the range of only 0.014 nmol/mg/min, which is 10-fold lower than those measured in E. coli (48), P. aeruginosa (52), and Clostridium (50). In B. subtilis, the Km for GABA transport is 37 μM (10). The apparent Kms measured here (l-arginine, 250 μM; GABA, 165 μM) are significantly higher than those described in other systems. The absolute levels of uptake of amino acids described in the present study point to low uptake levels as one possible impediment to utilization of the substrates for growth. In Salmonella enterica serovar Typhimurium for example, l-arginine transport severely limits l-arginine catabolism (31).
Our studies indicate that BCG exhibits unusual patterns of regulation of uptake of both GABA and l-arginine. The GABA utilization genes of B. subtilis, including the permease gene gabP, are regulated independently by nitrogen starvation and amino acid availability, and the organisms can use GABA as the sole nitrogen source (21). Klebsiella aerogenes can use GABA as both a carbon and nitrogen source, and GABA genes are induced by GABA in the medium (22). In E. coli, however, the GABA genes are expressed constitutively at low levels, and this species is incapable of growing on GABA as a sole source of either carbon or nitrogen (20). E. coli mutants with increased expression of the gab regulator (gabC) or the GABA permease (gabP) can grow on GABA (20). Our study shows that BCG does not grow with GABA as the sole carbon and nitrogen source, even at concentrations as high as 50 mM. Furthermore, incubation of cells with GABA (20 mM) as the sole carbon and nitrogen source in a minimal salts medium for 2 h or overnight did not increase the uptake of GABA by BCG. Accumulation of GABA is probably not sufficient to support the cells in the absence of any other carbon and nitrogen sources.
In B. subtilis, nitrogen starvation causes a 26-fold induction of expression of the GABA permease, encoded by the gabP gene. Unlike B. subtilis, the uptake of GABA by BCG was decreased even under conditions of overnight starvation. BCG transported GABA most efficiently when the cells were grown in Middlebrook medium containing appropriate carbon and nitrogen sources. Therefore, this permease is probably expressed when cells are in balanced growth in Middlebrook medium.
After incubation in exogenous l-arginine, arginine uptake by the mutant was dramatically increased to the levels seen in wild-type cells incubated in l-arginine. These results suggest that Rv052 may play a role in l-arginine efflux in wild-type cells: in the ΔRv052 mutant, this efflux activity may be absent, leading to greatly increased l-arginine uptake after incubation in exogenous l-arginine. Furthermore, these studies point to the presence of another l-arginine-responsive permease(s) expressed in the mutant. One candidate for this permease is a second rocE homolog discovered in the Sanger database (Rv2320c). Thus, as in the enterics (11, 53), l-arginine uptake is carried by more than one permease in BCG. Analysis of the regulation of expression of the two arginine transporters of M. tuberculosis and BCG (Rv0522 and Rv2320c) gene is under way.
It is puzzling that despite the array of l-arginine catabolic genes, both structural and regulatory, found in the M. tuberculosis databases, l-arginine and related amino acids are not utilized as sole carbon and nitrogen sources by BCG (this study) or M. tuberculosis (Erdman) (N. D. Connell, unpublished data). In addition, there are no previous reports of mycobacterial growth inhibition by any amino acids. Lyon et al. evaluated amino acid utilization by M. tuberculosis, and l-arginine was among the amino acids tested that were degraded, as measured by removal of l-arginine, supplied at 5 mM, from the culture supernatant (33). However, the study did not rely on the stringent test of utilization of these amino acids as the sole carbon or nitrogen source, since ammonium and glycerol were present in the media.
The mechanism by which exogenous l-arginine is growth inhibitory in BCG is not known. In S. enterica serovar Typhimurium, high concentrations of l-arginine (>5 mM) inhibit the enzyme ornithine carbamoyltransferase (ArgF) (1). High concentrations of l-arginine (15 mM) repress arginine biosynthetic enzymes 15–20 fold in Streptomyces coelicolor. In BCG, high concentrations of exogenous l-arginine may repress arginine biosynthesis. l-Arginine accumulation is probably not sufficient for growth but may be high enough to cause a complete repression of arginine biosynthesis. In many bacterial species, l-arginine as a corepressor binds to the ArgR repressor to repress l-arginine biosynthesis (34). Studies of ArgR regulation of l-arginine biosynthesis in BCG and M. tuberculosis are under way in our laboratory. Alternatively, since the enzymes involved in l-arginine synthesis are intimately involved in polyamine synthesis (24), excess l-arginine may lead to alterations in polyamine regulation in BCG.
Interestingly, l-arginine biosynthesis and transport have been shown in a number of pathogenic organisms to be important in intracellular metabolism. Early IVET studies in S. enterica serovar Typhimurium, revealed an l-arginine biosynthetic enzyme (carAB) (35). In Listeria monocytogenes, among the genes expressed preferentially in infected mammalian cells is arpJ, encoding an l-arginine transporter (30). No l-arginine biosynthetic mutants have been analyzed in mycobacterial host cell survival, although an l-arginine tRNA synthetase gene was specifically induced in M. marinum cells in infected macrophages (6). Thus, although shown in different bacterial systems, genes involved in l-arginine synthesis, transport, and incorporation into proteins are all increased during intracellular growth of bacteria. It is well established that l-arginine transport is stimulated in activated macrophages compared with resting macrophages, since l-arginine is absolutely required for the production of nitric oxide in the murine system (4, 8, 25, 29, 49). AS1 did not show a decrease in survival compared to wild-type BCG upon infection of J774.1 murine macrophages. It would be interesting to use multiple mutants of BCG in which more than one permease has been inactivated as probes for exploring possible interactions involving l-arginine between the host macrophage and the infecting mycobacterium.
ACKNOWLEDGMENTS
We thank Marty Pavelka for critical reading of the manuscript. We also thank the Molecular Resource Facility at the UMD/NJ Medical School for providing PCR primers and the sequencing facility.
This work was supported in part by Public Health Service Award 2R21AI34436-06A1 to N.D.C., by the Foundation of UMDNJ, and by the New Jersey Medical School National Tuberculosis Center.
REFERENCES
- 1.Abdelal A T H, Kennedy E H, Nainan O. Ornithine transcarbamylase from Salmonella typhimurium: purification, subunit composition, kinetic analysis and immunological cross-reactivity. J Bacteriol. 1977;129:1387–1396. doi: 10.1128/jb.129.3.1387-1396.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad S, Bhatnagar R K, Venkitasubramanian T A. Influence of carbon and nitrogen sources on arginine biosynthesis in Mycobacterium smegmatis ATCC 14468. Ann Microbiol (Paris) 1984;135B:137–146. doi: 10.1016/s0769-2609(84)80021-6. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad S, Bhatnagar R K, Venkitasubramanian T A. Ornithine transcarbamylase from Mycobacterium smegmatis ATCC 14468: purification, properties, and reaction mechanism. Biochem Cell Biol. 1986;64:1349–1355. doi: 10.1139/o86-177. [DOI] [PubMed] [Google Scholar]
- 4.Albina J E, Caldwell M D, Henry W L, Jr, Mills C D. Regulation of macrophage functions by l-arginine. J Exp Med. 1989;169:1021–1029. doi: 10.1084/jem.169.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasundaram D, Tyagi A K. Modulation of arginine decarboxylase activity from Mycobacterium smegmatis. Evidence for pyridoxal-5′-phosphate-mediated conformational changes in the enzyme. Eur J Biochem. 1989;183:339–345. doi: 10.1111/j.1432-1033.1989.tb14934.x. [DOI] [PubMed] [Google Scholar]
- 6.Barker L P, Brooks D M, Small P L. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol Microbiol. 1998;29:1167–1177. doi: 10.1046/j.1365-2958.1998.00996.x. [DOI] [PubMed] [Google Scholar]
- 7.Baumberg S, Harwood C R. Carbon and nitrogen repression of arginine catabolic enzymes in Bacillus subtilis. J Bacteriol. 1979;137:189–196. doi: 10.1128/jb.137.1.189-196.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baydoun A R, Bogle R G, Pearson J D, Mann G E. Arginine uptake and metabolism in cultured murine macrophages. Agents Actions. 1993;38:127–129. doi: 10.1007/BF01991160. [DOI] [PubMed] [Google Scholar]
- 9.Bhatt A, Green R, Coles R, Condon M, Connell N D. A mutant of Mycobacterium smegmatis defective in dipeptide transport. J Bacteriol. 1998;180:6773–6775. doi: 10.1128/jb.180.24.6773-6775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brechtel C, King S. 4-Aminobutyrate (GABA) transport from the amine-polyamine-choline superfamily: substrate specificity and ligand recognition profile of the 4-aminobutyrate permease from Bacillus subtilis. Biochem J. 1998;333:565–571. doi: 10.1042/bj3330565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celis R, Rosenfeld H J, Maas W K. Mutants of Escherichia coli K-12 defective in the transport of basic amino acids. J Bacteriol. 1973;116:619–626. doi: 10.1128/jb.116.2.619-626.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celis R T F. Chain-terminating mutants affecting a periplasmic binding protein involved in the active transport of arginine and ornithine in Escherichia coli. J Biol Chem. 1981;256:773–779. [PubMed] [Google Scholar]
- 13.Celis R T F. Mapping of two loci affecting the synthesis and structure of a periplasmic protein involved in arginine and ornithine transport in Escherichia coli K-12. J Bacteriol. 1984;151:1314–1319. doi: 10.1128/jb.151.3.1314-1319.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan J, Xing Y, Magliozzo R S, Bloom B R. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemens D L, Lee B Y, Horwitz M A. Purification, characterization, and genetic analysis of Mycobacterium tuberculosis urease, a potentially critical determinant of host-pathogen interaction. J Bacteriol. 1995;177:5644–5652. doi: 10.1128/jb.177.19.5644-5652.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 17.Connell N D. Mycobacterium: isolation, maintenance, transformation and mutant selection. Methods Cell Biol. 1994;45:107–125. doi: 10.1016/s0091-679x(08)61848-8. [DOI] [PubMed] [Google Scholar]
- 18.Connell N D, Nikaido H. Membrane permeability and transport. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection and control. Washington, D.C.: ASM Press; 1994. pp. 333–352. [Google Scholar]
- 19.Donnelly M I, Cooper R A. Succinic semialdehyde dehydrogenases of Escherichia coli. Eur J Biochem. 1981;113:555–561. doi: 10.1111/j.1432-1033.1981.tb05098.x. [DOI] [PubMed] [Google Scholar]
- 20.Dover S, Halpern Y S. Utilization of γ-aminobutyric acid by Escherichia coli K-12 mutants. J Bacteriol. 1972;109:835–843. doi: 10.1128/jb.109.2.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferson A E, Wray L V, Jr, Fisher S H. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich B, Magasanik B. Utilization of arginine by Klebsiella aerogenes. J Bacteriol. 1978;133:680–685. doi: 10.1128/jb.133.2.680-685.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardan R, Rapoport G, Debarbouille M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;23:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 24.Glansdorff N. Biosynthesis of arginine and polyamines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin A C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 25.Granger D L, Hibbs J B, Jr, Perfect J R, Durack D T. Metabolic fate of l-arginine in relation to microbiostatic capability of murine macrophages. J Clin Investig. 1990;85:264–273. doi: 10.1172/JCI114422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D. Studies of transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 28.Jarlier V, Nikaido H. Permeability barrier to hydrophilic solutes in Mycobacterium chelonae. J Bacteriol. 1990;172:1418–1423. doi: 10.1128/jb.172.3.1418-1423.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller R, Gehri R, Keist R, Huf E, Kayser F H. The interaction of macrophages and bacteria: a comparative study of the induction of tumoricidal activity and of reactive nitrogen intermediates. Cell Immunol. 1991;134:249–256. doi: 10.1016/0008-8749(91)90348-f. [DOI] [PubMed] [Google Scholar]
- 30.Klarsfeld A D, Goossens P L, Cossart P. Five Listeria monocytogenes gene preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE, and an arginine ABC transporter, argJ. Mol Microbiol. 1994;13:585–597. doi: 10.1111/j.1365-2958.1994.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 31.Kustu S G, McFarland N C, Hui S P, Esmon B, Ames G F-L. Nitrogen control in Salmonella typhimurium: coregulation of synthesis of glutamine synthetase and amino acid transport systems. J Bacteriol. 1979;138:218–234. doi: 10.1128/jb.138.1.218-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M H, Pascopella L, Jacobs W R J, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, BCG and M. tuberculosis. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon R H, Hall W H, Costas-Martinez C. Utilization of amino acids during growth of Mycobacterium tuberculosis in rotary cultures. Infect Immun. 1970;1:513–520. doi: 10.1128/iai.1.6.513-520.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maas W K. The arginine repressor of Escherichia coli. Microbiol Rev. 1994;58:631–640. doi: 10.1128/mr.58.4.631-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 36.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 37.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators that catalyze uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 38.Marinus M. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973;127:47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- 39.Marquis H, Bouwer H G A, Hinrichs D J, Portnoy D A. Intracytoplasmic growth and virulence of Listeria monocytogenes auxotrophic mutants. Infect Immun. 1993;61:3756–3760. doi: 10.1128/iai.61.9.3756-3760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masood R, Venkitasubramanian T A. Role of various carbon and nitrogen sources in the regulation of enzymes of pyrimidine biosynthesis in Mycobacterium smegmatis TMC 1546. Ann Inst Pasteur Microbiol. 1987;138:501–507. doi: 10.1016/0769-2609(87)90036-6. [DOI] [PubMed] [Google Scholar]
- 41.McNeil M R, Besra G S, Brennan P J. Chemistry of the mycobacterial cell wall. In: Rom W N, Garay S M, editors. Tuberculosis. New York, N.Y: Little, Brown & Co.; 1996. pp. 171–186. [Google Scholar]
- 42.Mountain A, Baumberg S. Map mutations of some mutations conferring resistance to arginine hydroxamate in Bacillus subtilis. Mol Gen Genet. 1980;178:691–701. doi: 10.1007/BF00337880. [DOI] [PubMed] [Google Scholar]
- 43.Nishijyo T, Park S, Lu C, Itoh Y, Abdelal A. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5559–5566. doi: 10.1128/jb.180.21.5559-5566.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad R, Kalra V K, Brodie A F. Different mechanisms of energy coupling for transport of various amino acids in cells of Mycobacterium phlei. J Biol Chem. 1976;251:2493–2498. [PubMed] [Google Scholar]
- 45.Ratledge C. Nutrition, growth and metabolism. In: Ratledge C, Stanford J, editors. The biology of the mycobacteria. I. London, United Kingdom: Academic Press; 1982. pp. 186–212. [Google Scholar]
- 46.Reyrat J M, Berthet F X, Gicquel B. The urease locus of Mycobacterium tuberculosis and its utilization for the demonstration of allelic exchange in Mycobacterium bovis bacillus Calmette-Guerin. Proc Natl Acad Sci USA. 1995;92:8768–8772. doi: 10.1073/pnas.92.19.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyrat J M, Lopez-Ramirez G, Ofredo C, Gicquel B, Winter N. Urease activity does not contribute dramatically to persistence of Mycobacterium bovis bacillus Calmette-Guerin. Infect Immun. 1996;64:3934–3936. doi: 10.1128/iai.64.9.3934-3936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosen B P. Basic amino acid transport in Escherichia coli: properties of canavanine-resistant mutants. J Bacteriol. 1973;116:627–635. doi: 10.1128/jb.116.2.627-635.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibazaki T, Fujiwara M, Sato H, Fujiwara K, Abe K, Bannai S. Relevance of the arginine transport activity to the nitric oxide synthesis in mouse peritoneal macrophages stimulated with bacterial lipopolysaccharide. Biochim Biophys Acta. 1996;1311:150–154. doi: 10.1016/0167-4889(95)00198-0. [DOI] [PubMed] [Google Scholar]
- 50.Speelmans G, Poolman G, Konings W. Amino acid transport in the thermophilic anaerobe Clostridium fervidus is driven by an electrochemical sodium gradient. J Bacteriol. 1993;175:2060–2066. doi: 10.1128/jb.175.7.2060-2066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennet L T, Bansal G P, Young J F, Lee M H, Hatfull G F, Snapper S B, Barletta R G, Jacobs J, R. W, Bloom B R. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 52.Verhoogt H J C, Smit H, Abee T, Gamper M, Driessen A J M, Haas D, Konings W N. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J Bacteriol. 1992;174:1568–1573. doi: 10.1128/jb.174.5.1568-1573.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G. A third periplasmic transport system for L-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol Microbiol. 1995;17:675–686. doi: 10.1111/j.1365-2958.1995.mmi_17040675.x. [DOI] [PubMed] [Google Scholar]
- 54.Yabu K. Amino acid transport in Mycobacterium smegmatis. J Bacteriol. 1970;102:6–13. doi: 10.1128/jb.102.1.6-13.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]