Abstract
The vast accumulation of protein structural data has now facilitated the observation of many different complexes in the PDB for the same protein. Therefore, a single protein complex is not sufficient to identify their interaction sites, especially for proteins with multiple binding states or different partners, such as hub proteins. PiSite is a database that provides protein–protein interaction sites at the residue level with consideration of multiple complexes at the same time, by mapping the binding sites of all complexes containing the same protein in the PDB. PiSite provides easy web interfaces with an interactive viewer working with typical web browsers, and the different binding modes can be checked visually. All of the information can also be downloaded for further analyses. In addition, PiSite provides a list of proteins with multiple binding partners and multiple binding states, as well as up-to-date statistics of protein–protein interfaces. PiSite is available at http://pisite.hgc.jp
INTRODUCTION
Protein–protein interactions are fundamental for proteins to exert their biological functions, and the molecular interactions of proteins can be understood by observing the 3D structure of their complexes. Therefore, many efforts have been made to solve complex structures experimentally, and a large number of structures are now available in the Protein Data Bank (PDB) (1). As a result of the vast accumulation of structural data from several genomic projects (2), we can now estimate the structural changes of proteins (3), the evolution of homo-oligomerization states (4,5) and the changes in protein–protein interactions (6) by analyzing all of the structures in the PDB simultaneously.
The structural characteristics of protein–protein interaction sites have been extensively studied (7,8), and the knowledge has been used for the prediction of binding sites (9–12). For the statistical analyses, one representative is usually selected for each group with similar amino acid sequences. However, some proteins, called hub proteins (13), interact with several kinds of different proteins, and thus one representative complex is not sufficient to describe all of the interfaces of a hub protein. For the identification of all of the binding sites of a hub protein, all of the complexes within the PDB should be considered at the same time.
Protein structures often consist of structural domains, and protein interactions can sometimes be interpreted as domain–domain interactions (14). Therefore, many databases to describe domain–domain interactions have been developed [3did (15), DIMA2 (15) and DOMINE (16)], and networks of domain interactions have been constructed. In these databases, large-scale interaction networks are the main focus, and the residue-level interactions are not described. On the other hand, for better understanding the molecular interactions of protein domains, SCOPPI (17) and iPFAM (18) provide residue-level information about the interacting domains, but the interacting residues are provided for each pair of interacting domains (iPfam) or a multiple alignment in the family with interface classification (SCOPPI), and thus it is difficult to observe several different binding modes of proteins with multiple partners. In addition, SCOPPI focuses on the diversity at the family level, while we would like to observe the multiple binding modes for individual proteins. In the similar way, PiBase (19) provides detailed physicochemical properties for all the protein complexes in the PDB as a correction of binary relationships.
In this article, we describe a new protein–protein interaction database, PiSite, based on the protein complexes in the PDB. We tried to provide a simple user interface to observe the ‘real’ binding sites, by simultaneously considering multiple binding states of individual proteins at the residue level, and not just for protein domains but for entire protein chains. PiSite also provides a list of ‘sociable proteins’, proteins with multiple binding states and multiple binding partners, which are considered as the key molecules in protein interaction networks. It should be noted that the sociable proteins are somewhat different from the so-called hub proteins, because the so-called hub proteins, which are usually defined as proteins with multiple binding partners in a protein interaction network obtained by large-scale experiments (20–23), are sometimes the subunits of a supermolecule (6). Furthermore, PiSite provides up-to-date statistics of protein interfaces. PiSite will be periodically updated every 3 months, using the most recent version of the PDB.
CONSTRUCTION METHOD
Data set
The current version of PiSite was constructed from the PDB entries as of July 2008. We did not use the asymmetric unit of the coordinates, but employed the biological units distributed by RCSB, to eliminate the crystallographic interfaces (ftp://ftp.rcsb.org/pub/pdb/data/biounit/coordinates). All protein chains with more than 30 residues were considered as proteins and we excluded the entries with >5.0 Å resolution and the models with only Cα coordinates. As a result, we selected 110 325 protein chains from the 51 482 PDB entries in our data set.
Mapping
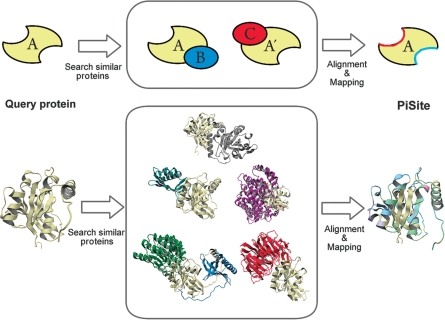
The binding sites that appeared in the PDB were gathered by mapping from all complexes to each protein chain (Fig. 1). For this purpose, at first, a similarity search by BLAST (24) against all protein chains in the data set was carried out for each protein chain. Ideally, the exact match of the amino acid sequence may be sufficient to find other complex structures in the PDB, but to enlarge the complex information and to avoid minor errors in sequence records due to missing residues and/or modifications, the proteins with sequence identity >90% and with coverage of the smaller protein >80% were used to select the entries for mapping. Then, mapping of the binding sites from one complex to a query chain was performed according to the BLAST alignment. The identification of the binding site residues was achieved by the distance criteria: when the minimum distance between the atoms in a residue pair was <4.0 Å, then the pair of residues was defined as contact residues. If the number of contacting residues between a pair of proteins was less than two, then the pair of protein chains was not used for mapping. It should be noted that we sometimes refer to the similar proteins used for the mapping as similar proteins for simplicity, but our focus is not to analyze the interfaces among the family members, in contrast to other protein–protein databases.
Figure 1.
An explanation of residue mapping. The upper panel shows a schematic representation of residue mapping, and the lower panel shows an example of the mapping by using 3D models. In the example, the GTP binding protein RAN (PDB: 1byu, chain A) was used. The gray, light blue, purple, green, blue and red chains were taken from 1byuB, 1a2kD, 1ibrB, 1k5gKL and 1l1mB, respectively (the four letter code indicates the PDB ID and the fifth letter means the chain ID).
Definition of the binding state
By selecting similar entries, as mentioned, we can enumerate all complexes containing the protein chain being considered, and can identify all of the binding partners that interact with the considering protein chain. The binding partners were grouped by sequence identity, and the number of groups was used as the number of binding partners shown in PiSite. Here, we regarded two proteins as being in the same group, if they have >30% sequence identity and >50% coverage of the smaller chain. Each binding state of a protein was defined as the combination and the number of binding partners appeared in the PDB. For example, when we consider the binding state of protein A with the complexes A–B and A–B–C in the PDB, then the number of binding states of protein A is two. If we have another complex A–B–B, then the number of binding states is three, because the number of binding partner B is different from that in the complex A–B. It should be noted that the similarities between binding sites are not considered explicitly in our approach, thus the number of binding state can be smaller than that of the binding modes, while we used all complexes for the residue mapping to get the comprehensive mapping.
The definitions of binding states and binding partners are virtually same as those used in Higurashi et al. (6), but we modified the original protocol to handle all PDB entries in PiSite, as described.
Definition of sociable proteins
We basically followed the sociable protein definition by Higurashi et al. (6). However, since automated processes are required in this study and we could not apply manual curation, we used a stricter definition. We defined proteins with three or more binding states and three or more binding partners as sociable proteins. We excluded proteins with more than 10 chains in a single PDB entry as supermolecules. In addition, we also excluded proteins with four or fewer similar proteins in the data set. This last condition is to ensure the reliability of sociable protein identifications. If the number of similar proteins is too small, then this definition may contain some errors.
CONTENTS OF PiSite
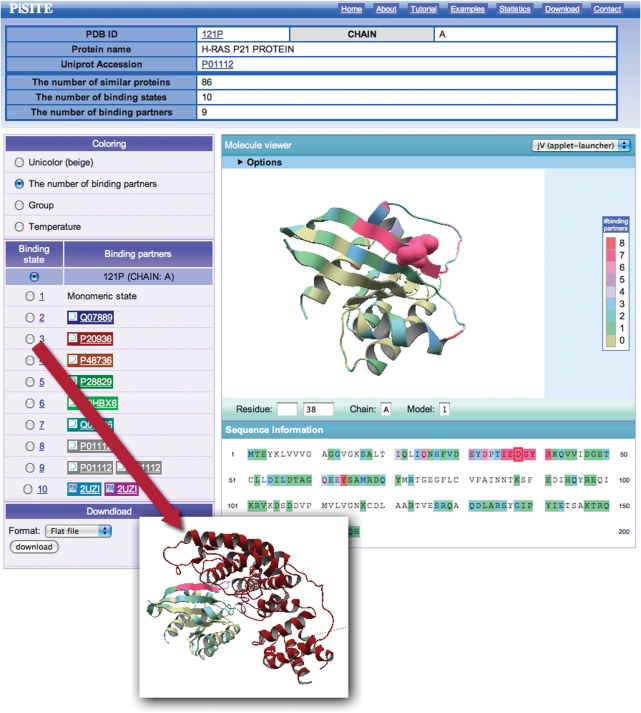
The main content of the PiSite is an interactive view of the multiple binding states, and we refer to the corresponding web page as the interaction viewer. The interaction viewer (Fig. 2) shows all binding states appearing in the PDB, and is prepared for each protein chain. It consists of four parts: title table including a link to UniprotKB (25), a molecule viewer, a binding state viewer and a download menu. The title table contains brief descriptions of the protein chain and the numbers of similar proteins, binding states and binding partners.
Figure 2.
An example of a PiSite entry. See the main text for details.
The molecule viewer shows the protein, colored according to the number of binding partners for each residue in default. The amino acid sequence is also colored according to the same procedure, and is shown just below the viewer. The position of each amino acid can be checked by clicking a residue in the sequence, which changes the specified residue into a CPK model (Fig. 2). Visualization of the molecule was done by jV [formally known as pdbjviewer (26)], and thus the view of the molecule can be interactively rotated and translated by mouse operations, and the residue position can be inspected by clicking the residue on the screen. More detailed options for jV are also available, by opening the option screen at the top of the molecule viewer (Fig. 2).
The binding state viewer shows a different binding state of the proteins, by communicating with the molecule viewer. By switching to a different binding state with the radio button, the complex structure is shown in the molecule viewer with the same color as the background of the partner name. It may be noteworthy that two or more protein names appear as binding partners as in the cases of the 9th and 10th binding states in Fig. 2, which means that the binding state contains three or more chains, including the chain under consideration. The names of the binding partners are taken from the DBREF record in the PDB entry. All of the data obtained after mapping the similar proteins can be downloaded in a flat file or XML format. The format is described in the download page.
OTHER CONTENTS
PiSite also provides a list of sociable proteins, which are proteins with multiple binding states and multiple binding partners. In our original paper, the number of sociable proteins was 86 (6), where only the structures with a resolution of 3.0 Å or better were selected from the PDB as of July 2006. In contrast, the number of nonredundant sociable proteins in the current release of PiSite is 102, where the redundancies were eliminated by selecting one representative from 30% identity sequence clusters. The difference arises mainly from the date of the PDB and the inclusion of NMR data in PiSite.
On the statistics page, we also show some statistical analyses of amino acid occurrence in the protein–protein interfaces based on the latest release of PiSite, which will be useful to develop statistical parameters to evaluate the likelihood of protein–protein interfaces.
All of the raw data are also available from the statistics page for further analyses.
ACCESS TO EACH ENTRY, WITH AN EXAMPLE
PiSite provides two different ways to access each entry. The first way is access from a compiled list. PiSite provides a list of representative sociable proteins and supermolecules, as mentioned above. These lists contain links for the entries and the user can access them by clicking the links. The other way is to search by sequence or keywords. Both sequence and keyword searches are available, through the search form on the top page. A sequence search requires amino acid sequence of a protein chain as an input and is performed by using BLAST. A more general search is by keywords. If the user wants to check the PiSite entry for Ras p21 proteins, for example, then the user can search ‘RAS p21’ as a keyword. As a result, the search results, including a moleculer description of the _target and information about the number of binding states and partners are obtained. Each PiSite entry can be accessed from links in a search result page. In this case, the user can access the PiSite entry of the hRas P21 protein (PDBID: 121p, chain A) by clicking the link of the top hit of the search results (Fig. 2). If the PDB ID of a _target protein is already known, then rapid search can be available selecting the ‘search by PDB ID’ radio button. The interaction viewer shows that the protein has 10 different binding states and 9 different binding partners. The molecule viewer shows the number of binding partners of each residue, and the residues contacting many partners are colored magenta. Interestingly, this magenta-colored region, including residues 32–42, corresponds to the effector region of a Ras protein. This region of a Ras protein is known as a hot spot and has been identified as a neutralizing epitope of hRas (27). More mapping result details can be obtained from the download menu.
FUNDING
Institute for Bioinformatics Research and Development; Japan Science and Technology Corporation (BIRD-JST); Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.K.). Funding for open access charge: Annual budget for Human Genome Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 3.Koike R, Amemiya T, Ota M, Kidera A. Protein structural change upon ligand binding correlates with enzymatic reaction mechanism. J. Mol. Biol. 2008;379:397–401. doi: 10.1016/j.jmb.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: a structural classification of protein complexes. PLoS Comp. Biol. 2006;2:e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higurashi M, Ishida T, Kinoshita K. Identification of transient hub proteins and the possible structural basis for their multiple interactions. Protein Sci. 2008;17:72–78. doi: 10.1110/ps.073196308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, Thornton JM. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 1995;63:31–65. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 8.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of Protein-protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 9.Huang B, Schroeder M. Using protein binding site prediction to improve protein docking. Gene. 2008;422:14–21. doi: 10.1016/j.gene.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H-X, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007;23:2203–2209. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- 11.Valdar WS, Thornton JM. Protein-protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001;42:108–124. [PubMed] [Google Scholar]
- 12.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 13.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 14.Kim W, Bolser D, Park J. Large-scale co-evolution analysis of protein structural interlogues using the global protein structural interactome map (PSIMAP) Bioinformatics. 2004;20:1138–1150. doi: 10.1093/bioinformatics/bth053. [DOI] [PubMed] [Google Scholar]
- 15.Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acid Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghavachari B, Tasneem A, Przytycka TM, Jothi R. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2007;36:656–661. doi: 10.1093/nar/gkm761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn R, Marshall M, Bateman A. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 19.Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 20.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 22.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 23.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.UnitProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 27.Sigal IS, Gibbs JB, D’Alonzo JS, Scolnick EM. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc. Natl Acad. Sci. USA. 1986;83:4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]