Abstract
We report a mouse model of multiple osteochondromas (MO), an autosomal dominant disease in humans, also known as multiple hereditary exostoses (MHE or HME) and characterized by the formation of cartilage-capped osseous growths projecting from the metaphyses of endochondral bones. The pathogenesis of these osteochondromas has remained unclear. Mice heterozygous for Ext1 or Ext2, modeling the human genotypes that cause MO, occasionally develop solitary osteochondroma-like structures on ribs [Lin et al. (2000) Dev Biol 224(2):299–311; Stickens et al. (2005) Development 132(22):5055–5068]. Rather than model the germ-line genotype, we modeled the chimeric tissue genotype of somatic loss of heterozygosity (LOH), by conditionally inactivating Ext1 via head-to-head loxP sites and temporally controlled Cre-recombinase in chondrocytes. These mice faithfully recapitulate the human phenotype of multiple metaphyseal osteochondromas. We also confirm homozygous disruption of Ext1 in osteochondroma chondrocytes and their origin in proliferating physeal chondrocytes. These results explain prior modeling failures with the necessity for somatic LOH in a developmentally regulated cell type.
Keywords: osteochondroma, exostosis, loss of heterozygosity, growth plate, bone
Multiple osteochondromas (MO), alternatively called diaphyseal aclasis, osteochondromatosis, and multiple hereditary exostoses (MHE or HME), is an autosomal dominant disorder characterized by shortened long bones and multiple cartilage-capped bony projections from the metaphyses of endochondral bones that develop during statural growth (1). In 0.5–3% of patients with MO, an osteochondroma eventually transforms into a chondrosarcoma (2). MO has been linked to loss-of-function mutations in the EXT1 (8q24.1) (3) and EXT2 (11p11-p12) (4) genes (5).
EXT1 and EXT2 encode ubiquitously expressed type II transmembrane glycosyltransferases (6), which localize to the endoplasmic reticulum and Golgi complex (7). The two genes act in hetero-oligomeric complexes performing the N-acetylglucosamine and D-glucuronic acid transferase activities required for the synthesis of heparan sulfate (HS) chains on proteoglycans (PGs) (8). Besides their expected structural function, HSPGs have been shown to regulate ligand distribution and/or receptor binding of many signaling systems including the hedgehog, Wnt, and fibroblast growth factor families (9).
Whereas many theories on the pathogenesis of osteochondromas in the setting of MO have been proposed, each hinges on two central questions. (i) Is the cellular origin of an osteochondroma a chondrocyte of the growth plate or a cell from the juxtaposed perichondrium/periosteum? (ii) Is an osteochondroma the result of haploinsufficiency-dependent misregulation of signals controlling growth plate maturation or of clonally occurring second mutations in the EXT genes (10) as has been shown for other genetically inherited tumor susceptibililty syndromes (11)? Second mutations in or cytogenetic loss of the wild-type allele of EXT1 or EXT2 have been detected, but only in a subset of osteochondromas, leaving the pathogenetic role of loss of heterozygosity (LOH) unestablished (12–16).
In mice, deletion of Ext1 or Ext2 results in a lack of HS biosynthesis; homozygous embryos die between E6.5 and E9.5 due to failure in mesoderm formation. Mice heterozygous for loss of Ext1 or Ext2 survive and are fertile. Small, solitary, osteochondroma-like structures on ribs have been detected in 30% of heterozygous Ext2+/− mice. However, unlike humans with MO, heterozygosity for neither Ext1 nor Ext2 disruption has led to shortened long bones or frequent development of larger osteochondromas on other endochondral bones (17, 18). Here we demonstrate that clonal, homozygous inactivation of Ext1 in chondrocytes at low prevalence results in frequent osteochondromas on the apendicular skeleton, mimicking the human MO phenotype.
Results
Strategy to Model Loss of Heterozygosity.
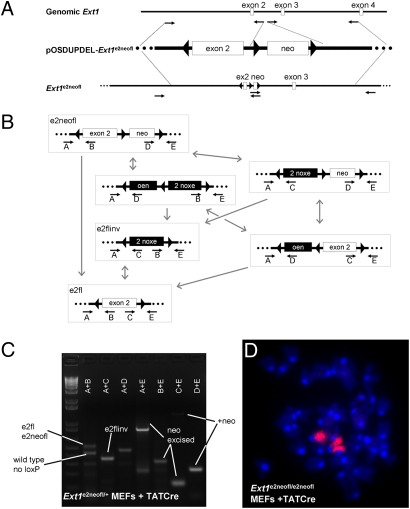
Exon 2 of Ext1 codes for highly conserved amino acids of the catalytic center (100% amino acid homology between mouse and human). Missense mutations in exon 2, which lead to MO in humans, have been demonstrated to completely abrogate the enzymatic function of EXT1 in vitro (7, 19). To investigate if low-prevalence clonal inactivation of Ext1 would result in osteochondroma formation in mice, we generated mice carrying an inducible loss-of-function allele of Ext1 (Ext1e2neofl), in which exon 2 was flanked by head-to-head loxP sites (Fig. 1A). Both heterozygous and homozygous carriers of the Ext1e2neofl allele were healthy, displaying no obvious phenotype.
Fig. 1.
Generation and characterization of head-to-head floxed Ext1. (A) Homology arms procured by long-range PCR (small arrows: PCR primers) from genomic Ext1 DNA (white boxes, exons; black lines, introns) were subcloned into pOSDUPDEL to either side of the neomycin resistance cassette (neo) flanked by forward orientation loxP sites (triangles). An inverted loxP site was introduced upstream of exon 2. _targeting was tested with long-range PCR. (B) Cre-mediated recombinations (gray arrows) produce a distribution of products (black boxes: inverted sequence) from any given state, including entire construct inversion, exon 2 inversion, or neo excision. After neo excision (e2fl or e2flinv), only exon 2 inversion and reversion are possible. Each of the recombination states is detectable by the primer pairs listed below it (small arrows). (C) PCR using template DNA isolated from Ext1e2neofl/+ MEFs after treatment with TATCre demonstrates presence of all recombination states. (D) Ext1e2neofl/e2neofl MEFs after TATCre demonstrated the normal diploid murine karyotype of 40 chromosomes counterstained with DAPI including 2 stained with FISH chromosome 15 paint (magenta).
Although prolonged exposure of head-to-head loxP sites to Cre-recombinase has been demonstrated to cause chromosomal loss and apoptosis due to interchromatid translocations (20–23), limited exposure to Cre-recombinase yields a distribution of forward and inverted flanked fragments (24). Homozygosity for a head-to-head flanked allele was therefore hypothezised to generate chimeric tissues containing a low prevalence of inversion of both alleles.
To determine if the previously described chromosomal loss and apoptosis from inverted loxP sites was applicable to our allele, we crossed homozygous Ext1e2neofl/e2neofl mutants to mice carrying Cre-recombinase under the control of the collagen IIa1 promoter (Col2-Cre) (25). No double-heterozygous Col2-cre;Ext1e2neofl/+ offspring (0 of 113) were produced. Similarly, Ext1e2neofl/e2neofl mutants crossed to mice expressing Cre-recombinase from the Hprt locus produced only very small litters. No double-heterozygous Hprt-Cre;Ext1e2neofl/+ mice were retrieved even at E11.5 (zero of four matings) (Table 1).
Table 1.
Genotype–phenotype correlation among Ext1 mutants
| Genotype | Cre induction | Age | Analysis (sample no.) | Phenotype |
| Ext1e2neofl/e2neofl | NA (rec’d dox) | 6–10 weeks | Histology: knees, ribs (5) | No osteochondromas |
| Ext1e2fl/e2fl | NA (rec’d dox) | 4–15weeks | Histology: knees, ribs (15) | No osteochondromas |
| Ext1e2flinv/e2flinv | NA | E7.0 | Histology and 10E4 IHC (4 of 10 pups) | Failed gastrulation and absent embryonic HS |
| Hprt-Cre; Ext1e2neofl/+ | Continuous | Weaning | Genotyping (0 from 3 litters, 9 pups) | Prenatal mortality |
| Hprt-Cre; Ext1e2neofl/+ | Continuous | E11.5 | Genotyping (0 from 1 litter, 4 pups) | Pre-E11.5 mortality |
| Col2-Cre; Ext1e2neofl/+ | Continuous | Weaning | Genotyping (0 from 113 pups) | Prenatal mortality |
| Col2-rtTA-Cre; Ext1e2neofl/+ | Week 2 doxycycline | 6–10 weeks | Histology: knees and ribs (7) | No osteochondromas. |
| Col2-rtTA-Cre; Ext1e2neofl/e2neofl | Week 2 doxycycline | 6–10 weeks | Histology: knees and ribs (17), μCT (10) | 100% osteochondromas; epiphyseal enchondromas |
| Col2-rtTA-Cre; Ext1e2fl/e2fl | Week 2 doxycycline | 4–15 weeks | Histology of knees (28) | 100% osteochondromas; epiphyseal enchondromas |
| Osx-CreERT; Ext1e2neofl/e2neofl | P8 tamoxifen | 9 weeks | Histology: knees plus μCT (4) | No osteochondromas; epiphyseal enchondromas |
To test if chromosomal loss and apoptosis could be avoided by limited exposures to Cre-recombinase, we exposed E14.5 mouse embryonic fibroblasts (MEFs) isolated from heterozygous Ext1e2neofl/+ and homozygous Ext1e2neofl/e2neofl mutants to TATCre (26). The cells remained healthy over at least five passages and had normal cytogenetic profiles on standard chromosomal spreads. Fluorescent in situ hybridization (FISH) with a chromosome 15 paint demonstrated the presence of the expected complement of two chromosomes 15 in homozygous Ext1e2neofl/e2neofl cells after TATCre exposure (Fig. 1D). Genotyping of the Ext1 allele detected all possible Cre-mediated recombination products predictable from the design of the _targeting construct (Fig. 1 B and C).
Accordingly, introduction of a transgenic Cre-recombinase under the control of a doxycycline-inducible Col2 promoter (Col2-rtTA-Cre) (27) generated viable Col2-rtTA-Cre;Ext1e2neofl/+ and Col2-rtTA-Cre;Ext1e2neofl/e2neofl double mutants. Furthermore, induction of Cre activity during gestation after two matings between Col2-rtTA-Cre;Ext1e2neofl/+ mice resulted in one Col2-rtTA-Cre;Ext1e2neofl/+ and one Col2-rtTA-Cre;Ext1e2neofl/e2neofl offspring. PCR-based genotyping of tail and other cartilaginous tissues revealed the presence of all predictable recombinations. Sequencing of the individual PCR products confirmed the expected identity of the rearranged fragments.
Exon 2 Inversion Disrupts Ext1 Function.
To simplify recombination products of the allele, we crossed Ext1e2neofl/e2neofl males to Prx1-cre females (28), which express Cre-recombinase in the female germ line. By PCR we screened for mutants that had lost the neo cassette and retained exon 2 in either wild-type (Ext1e2fl) or inverted (Ext1e2flinv) orientation (Fig. 1B). Stable germ-line transmission of the generated alleles was confirmed by crossing founders (F0) to C57B6 females. F1 males that produced offspring (F2) consistently carrying the allele in the expected orientation were used for further breeding. Both heterozygous Ext1e2fl/+ and Ext1e2flinv/+ mice were born at Mendelian ratios.
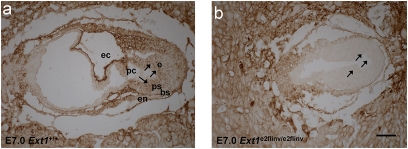
To verify that exon 2 inversion generates a functionally null allele, we isolated embryos of Ext1e2flinv/+ matings at E7.0. We detected 4 of 10 embryos that were small and had failed to undergo gastrulation (Fig. 2B), similar to the phenotypes described for Ext1−/− and Ext2−/− mutants (17, 18). PCR analysis of microdissected embryonic tissue confirmed that these were homozygous Ext1e2flinv/e2flinv mutants. To further support the inactivity of the inverted allele we investigated the production of HS by immunohistochemistry using the 10E4 (Seikagaku) antibody. In contrast to wild-type and heterozygous embryos, which showed solid staining in both the embryonic and extraembryonic tissues and basal membranes, we detected HS only in the placenta, but not in the basal membranes of the Ext1e2flinv/e2flinv embryos proper (Fig. 2).
Fig. 2.
Exon 2 inversion disrupts the function of Ext1. Immunohistochemistry of E7.0 frontal sections of wild-type (A) and Ext1e2flinv/e2flinv (B) embryos with 10E4 antibody demonstrated failing gastrulation and reduced HS in the embryonic tissue (arrows) of Ext1e2flinv/e2flinv, whereas placental tissues showed intensive staining for HS (ec, exocoelomic cavity; pc, proamniotic cavity; e, ectoderm; ps, primitive streak; bs, basement membrane; en, endoderm). (Scale bar, 50 μm.)
Postnatal Inactivation of Ext1 Generates Osteochondromas.
To investigate whether clonal inactivation of Ext1 function postnatally would lead to exostosis formation in mice, we induced expression of Cre-recombinase in chondrocytes from postnatal day 8 (P8) to P15 by administration of doxycycline in the drinking water.
Analysis of hematoxylin and eosin (H&E) and Safranin-O stained serial sections of knees and rib cages did not detect osteochondromas in 4-, 6-, or 10-week-old mutants, which lacked Cre-recombinase and/or homozygosity for the Ext1e2fl or Ext1e2neofl allele (Table 1). In contrast, Col2-rtTA-Cre;Ext1e2neofl/e2neofl and Col2-rtTA-Cre;Ext1e2fl/e2fl mice formed multiple osteochondromas with 100% penetrance after induction with doxycyline (Table 1 and Fig. 3). No differences could be detected in mutants with and without the neo cassette, excluding a nonspecific effect of the cassette itself on phenotype (29). To more precisely specify the induction time, we switched to administration of doxycycline by i.p. injection to the lactating dam. One i.p. administration of doxycycline to the mother at P8 was sufficient to induce osteochondromas with 100% frequency. No difference in osteochondroma morphology was observed between the two administration strategies.
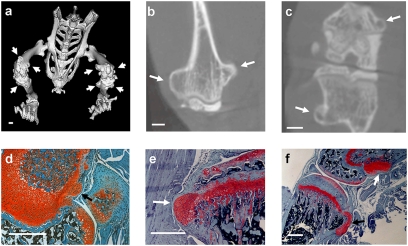
Fig. 3.
Postnatal inactivation of Ext1 in chondrocytes generates osteochondromas. (A) Three-dimensional microCT rendering of a 10-week-old Col2-rtTA-Cre;Ext1e2neofl/e2neofl mouse after doxycyline treatment shows osteochondromas on the distal femur and proximal tibia (arrows). Coronal plane (B) and axial plane (C) reconstructions of the distal femur confirm cortical and medullary continuity of the osteochondromas, matching radiographic definitions for human diagnosis. Safranin-O stained sections from the knees of 4- (D), 6- (E), and 10-week-old (F) Col2-rtTA-Cre;Ext1e2neofl/e2neofl mice show early, middle, and fully developed osteochondromas, respectively, and both pedunculated (F, black arrow), and sessile (F, white arrow) morphologies.
As in human patients, the induced osteochondromas developed on most endochondral bones and were mainly located near the major growth centers. Both described osteochondroma morphologies, pedunculated and sessile, were observed (Fig. 3F). Planar reconstructions from microCT imaging revealed cortical and medullary continuity of each osteochondroma with the host bone, consistent with the radiographic diagnostic criteria in humans (Fig. 3 B and C).
To investigate the tissue composition of the osteochondromas, we analyzed sections through knees and rib cages at 4, 5, 6, and 8 weeks of age on a morphological and molecular level. In every mouse ≥6 weeks of age, safranin-O and H&E staining demonstrated cartilage-capped bone protuberances with cortical and medullary continuity and morphologic recapitulation of the physeal zones (Fig. 3). The morphological observations were supported by in situ hybridization analysis (Fig. S1), revealing a region of Col-II expression in cells at the peripheral cap of the osteochondroma and in columnar chondrocytes, which was followed by Ihh-expressing prehypertrophic chondrocytes and Col-X-expressing hypertrophic chondrocytes. As in osteochondromas arising in human MO patients, the osteochondromas induced in Col2-rtTA-Cre;Ext1e2fl/e2fl mice mimic the organization of the growth plate on a molecular and morphological level.
Osteochondromas Originate in Proliferating Chondrocytes.
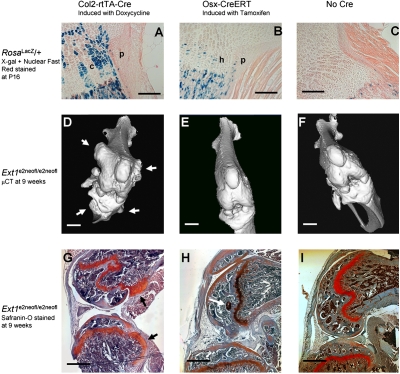
The cellular origin of the osteochondroma has been disputed to be either a peripheral physeal chondrocyte or a cell in the adjacent perichondrium/periosteum. As the Col2 promoter has previously been shown to drive expression in both tissues embryonically (30), we analyzed Col2-rtTA-Cre;Rosa26LacZ/+ mutants that had received doxycycline by drinking water at P8–P12. β-Galactosidase (β-gal) staining at P16 demonstrated expression of the allele in ≈50% of proliferating growth plate chondrocytes and rarely in cells of the periosteum and primary spongiosa (n = 5). Although showing infrequent Cre-mediated recombination, these periosteal cells could not be thus excluded as the origin for osteochondromas (Fig. 4A).
Fig. 4.
The proliferating chondrocyte is the cell of origin for an osteochondroma. (A) β-Gal staining of a P16 proximal tibial physis from Col2-rtTA-Cre;Rosa26LacZ/+ after P8–12 doxycyline by drinking water shows abundant staining of proliferating chondrocytes (C) with minimal staining of the inner layer of cells in the perichondrium/periosteum (P). (B) Osx-CreERT;Rosa26LacZ/+ mice after P8 injection of tamoxifen have more robust staining of the inner layer of perichondrium/periosteum (P), with additional staining of hypertrophic chondrocytes (H), but not proliferating chondrocytes. (C) Rosa26LacZ/+ mice without Cre-recombinase demonstrate only mild background staining of primary spongiosal osteoblasts. Three-dimensional knee renderings of microCTs from 9-week-old Col2-rtTA-Cre;Ext1e2neofl/e2neofl (D), Osx-CreERT;Ext1e2neofl/e2neofl (E), and Ext1e2neofl/e2neofl (F) mice demonstrate the presence of osteochondromas (arrows) only on the Col2-rtTA-Cre mutants. Safranin-O staining from the same groups confirms the presence (G, black arrows) and absence (H and I) of osteochondromas, but also the presence of a small epiphyseal enchondroma (H, white arrow) in the Osx-CreERT mutant, likely from its hypertrophic chondrocyte Cre activity. (Scale bars: A–C, 100 μm; D–I, 500 μm; background artifact has been removed from panel 4G).
We next introduced a tamoxifen-inducible Cre-recombinase under the control of the Osterix promoter (Osx-CreERT) (31) to Rosa26LacZ/+ and Ext1e2neofl/e2neofl mutants. β-Gal staining of P16 knees from Osx-CreERT;Rosa26LacZ/+ mice that had received i.p. tamoxifen at P8 revealed expression in the osteoblasts of the perichondrium/periosteum and the primary spongiosa (n = 3). Expression was also detected in hypertrophic chondrocytes of the physis, but was excluded from the proliferative zone (Fig. 4B). MicroCT analysis of the lower extremities of Osx-CreERT;Ext1e2neofl/e2neofl mice induced with tamoxifen at P8 did not detect any signs of osteochondroma development at 9 weeks of age (n = 4) (Fig. 4E). PCR on genomic DNA derived from bone tissues confirmed that recombination had taken place. Furthermore, safranin-O staining of serial sections of knees demonstrated mild aberrations in hypertrophic chondrocytes, but no osteochondromas (Fig. 4H), strongly indicating that the likely origin of the osteochondroma is a proliferating growth plate chondrocyte, the only localized cell type expressing Col2-rtTA-Cre, but not Osx-CreERT.
To confirm the proliferating chondrocyte as the cell of origin, we investigated sections through knees of doxycycline-induced Col2-rtTA-Cre;Ext1e2fl/e2fl mutants at 4 and 5 weeks of age. Small clusters of cells were noted, disrupting the columnar organization at the periphery of the growth plate. At slightly later stages, expanding peripheral clones of chondrocytes were found at the level of the primary spongiosa, often still in continuity with the proliferating zone of the physis. In situ hybridization with chondrocyte-specific markers revealed expression of Col-II and Ucma, but not Ihh and Col-X, confirming an early differentiation state in the chondrocytes of these early osteochondromas (Fig. S2).
Homozygous Loss of Ext1 Is Required for Osteochondromagenesis.
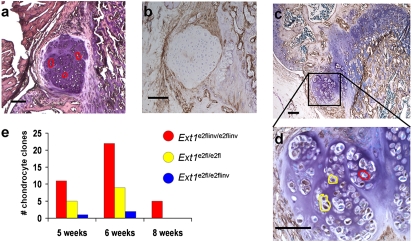
To confirm that osteochondromas are generated from homozygous mutant cells, we isolated clones of chondrocytes from sections of Col2-rtTA-Cre;Ext1e2fl/e2fl exostoses at different stages of development by laser capture microdissection (LCM) (Fig. S3). Genomic PCR using nested primer pairs to detect the Ext1e2fl and the Ext1e2flinv allele, respectively, revealed a mixture of all possible genotypes. About 70% of captured cell clusters produced neither the forward orientation nor the inverted PCR product, fitting the technical success rate from other studies using sections of paraformaldehyde-fixed, paraffin-embedded specimens for LCM (32, 33). Morphologically distinct, small clones of cells frequently displayed a homozygous Ext1e2fl/e2fl or Ext1e2flinv/e2flinv genotype, indicating that the clustered appearance reflects a genetically clonal origin and that recombination is rare after doxycycline clearance. Of 55 clones successfully analyzed, 38 were homozygous for the inverted Ext1e2flinv/e2flinv allele, 14 homozygous for the Ext1e2fl/e2fl allele, and 3 heterozygous Ext1e2fl/e2flinv (Fig. 5).
Fig. 5.
Osteochondroma chondrocytes demonstrate clonal inversion of exon 2 of Ext1. (A and B) Immunohistochemistry with 10E4 antibody in parallel sections showed that HS negativity correlated with homozygosity for the inverted allele (red-encircled clones). HS-positive chondrocytes (D, enlarged from C) demonstrated the Ext1e2fl/e2fl (yellow-encircled clones) genotype. (E) The distribution of 55 LCM-identified clone genotypes from osteochondroma chondrocytes demonstrates a strong majority of Ext1e2flinv/e2flinv genotypes at all ages. (Scale bars, 100 μm.)
To understand the spatial distribution of genotypes within the osteochondromas, we tested parallel sections with LCM genotyping and 10E4 immunohistochemistry, confirming that HS-negative areas are consistently Ext1e2flinv/e2flinv and that osteochondromas containing Ext1e2fl/e2fl chondrocytes displayed small, discrete regions of HS synthesis (Fig. 5). Screening of 10 osteochondromas with 10E4 immunohistochemistry demonstrated 6 with no detectable HS in the cap chondrocytes and 4 with a small minority of cells staining for HS, confirming that clonal dysfunction of Ext1 is present in the majority of chondrocytes in all osteochondromas, but that cells with functional Ext1 can be integrated into the mutant tissue during osteochondromagenesis.
Discussion
Phenotypic expression in mouse models of human genetic diseases that depend on somatic LOH is usually not detectable until late in the mouse life cycle (34, 35). The stochastic likelihood of achieving such LOH in a developmentally limited, short-lived tissue, such as proliferating physeal chondrocytes, is very low. Furthermore, somatic LOH is not well modeled by high-density disruption of a gene in the tissue of interest, as would be typical of a traditional conditional knockout strategy.
To mimic the heritable form of human osteochondromagenesis, MO, we have generated mice in which head-to-head loxP sites flank the second exon of Ext1. We demonstrate that limited duration of Cre-mediated recombination in chondrocytes leads to clonal inactivation of the Ext1 gene with low prevalence. In contrast to ubiquitous deletion of one or both copies of Ext1, such clonal deletions generate exostoses with high penetrance, strongly indicating that LOH is the inductive event for osteochondromas in humans. Although conflicting data have been published on the genetic status of human osteochondroma chondrocytes (2), somatic loss of the wild-type EXT1 or EXT2 allele has been identified in many hereditary and even some sporadic osteochondromas (36) and may have been masked in others by the occasional integration of wild-type chondrocytes into the osteochondroma cap, as seen in a minority of murine-induced osteochondromas. There is corroborating evidence for LOH as a mechanism from Ext2 null zebrafish that have disorganized rather than ordered columnar chondrocyte proliferation (37). In the mice, immunohistochemistry against HS proved a reliable surrogate for LCM genotyping and strongly supported the absence of Ext1 function in the majority of chondrocytes in all osteochondromas. Interestingly, reduced HS levels have been found in most human osteochondromas independent of genetically identified LOH (38, 39).
Investigation of the osteochondroma cellular origin has also been challenging in humans, as only late-stage osteochondromas are routinely surgically retrieved for study. These demonstrate continuity with the cortical and medullary bone, suggesting an at least temporary defect in the bone collar during osteochondromagenesis. Direct injury to the perichondrial ossification groove has induced solitary osteochondroma-like structures in prior animal models (40). However, although EXT1- or EXT2-null chondrocytes have been identified in the cartilaginous caps of osteochondromas, the perichondrium over the same has shown normal EXT gene expression when investigated (36). Further, a temporary defect in the bone collar might alternatively be generated by altered signaling to the perichondrium, rather than a cell-intrinsic defect there. Peripheral displacement of the early osteochondroma chondrocytes from the growth plate may push the overlying perichondrium away from the osteogenic signals from the physis or provide alternate, nonosteogenic, paracrine signals (Fig. 6). Although the disparate results of deleting Ext1 function in Osx- versus Col2-expressing cells in no way exclude a contribution of the perichondrial cells to osteochondromagenesis, they strongly indicate that the original molecular event, the clonal inactivation of HS synthesis, takes place in a growth plate chondrocyte.
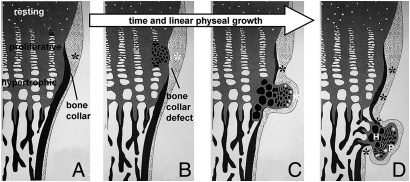
Fig. 6.
Osteochondromagenesis. (A) An early proliferative chondrocyte with homozygous disruption of Ext1 has no HS on its cell surface or in the immediate extracellular matrix (black clone of cells). (B) This loss of HS delays the clone’s differentiation and gives it a proliferative advantage. The immediately adjacent perichondrium fails to differentiate into osteoblasts and form a bone collar (white asterisk) due to blocked osteogenic signals from the physis, physical displacement away from the osteogenic signals from the physis, or receipt of alternate signals from the less-differentiated chondrocytes of the early osteochondroma. (C) Once the clone has progressed past the hypertrophic zone, the physis-flanking perichondrium receives normal osteogenic signals (black asterisks) again and reforms the bone collar in the wake of the osteochondroma. The osteochondroma may or may not have brought along cells of normal genotype (white cells, C and D) from the physis. (D) The fully developed osteochondroma, once completely independent from direct physeal signaling, begins to organize itself loosely into the proliferative (P) and hypertrophic (H) zones of a growth plate, as well as form its own primary spongiosa and bone collar, each in continuity with the host bone.
Although our data settle important controversies, many questions remain unanswered. What happens to a central clone of chondrocytes lacking Ext1 function? Although we did observe rare epiphyseal enchondromas, we did not detect them in the metaphysis. Also, what signals are altered by the clonal Ext1 dysfunction that gives rise to osteochondromas? Although only Ihh has been demonstrated to be affected by Ext1 dysfunction specifically in chondrocytes (41), the diffusion gradients and ligand-receptor interactions of other pathways important to physeal development have been shown to be sensitive to HS levels in other tissues (9). We did not find obviously aberrant expression of Ihh or Ptch1 in early osteochondromas. Further investigation of Ihh and other signaling systems will be required to elucidate the contribution of individual signal disturbances from HS absence.
Materials and Methods
Transgenic Mice.
A total of 3.6 kb of genomic DNA including exon 2 of Ext1 (42) [chromosome (chr) 15: 53,135,004–53,138,589] was amplified by PCR from the murine embryonic stem (ES) cell line 129 SvJ and cloned into the XhoI site of pMyr (Stratagene). An inverted loxP site was introduced upstream to exon 2 into a SacI site generated by site-directed mutagenesis using Quick Change (Stratagene). The fragment was then subcloned into the XhoI site of pOSDUPDEL upstream of the forward-facing loxP-flanked neomycin resistance (Neo) cassette. A total of 5.3 kb of DNA (chr 15: 53,138,682–53,144,025) beginning in intron 2 were amplified by PCR and subcloned into the EcoRI site downstream from the floxed Neo cassette in pOSDUPDEL (pOSDUPDEL-Ext1e2neofl; Fig. 1A).
The linearized _targeting vector was electroporated into R1 ES cells (129 × 1/SvJ3 129S1/Sv). G418-resistant clones were screened by long-range PCR to identify Ext1-_targeted ES cell lines. Ends of PCR products were sequenced to confirm identity. One positive cell line was injected into blastocysts. Germ-line transmission and further genotyping were initially confirmed by long-range PCR from tail-tip-derived DNA. Routine genotyping was done with primers A and B of a panel of primers used to detect recombination (Fig. 1 B and C).
Rosa26LacZ (43), Col2-rtTA-Cre (27), Hprt-Cre (44), Col2-Cre (25), and Osx-CreERT (31) mice were genotyped as described. In addition, presence or absence of Cre-recombinase was confirmed by PCR in each mouse.
Expression of Cre-recombinase in Col2-rtTA-Cre mice was induced either by administration of 4 mg/mL doxycycline in 5% sucrose through the drinking water of the lactating mother at P8 for 8 days or by i.p. injection of doxycycline (80 μg/g body mass) in PBS into lactating mothers at P8. Nuclear translocation of CreERT was induced in Osx-CreERT mice by i.p. administration of tamoxifen (0.2 mg/g body mass) in corn oil directly to the mutants.
Cell Culture.
E14.5 embryos from Ext1e2neofl/+ matings were harvested, disemboweled, dissociated in trypsin, and plated in Dulbecco’s modified Eagle medium (DMEM) with 10% FBS. Nonadherent cells were discarded. Washed cells were treated with TATCre (4 mM in DMEM) for 3 h before adding serum-containing media. For cytogenetics, cultures were arrested with methotrexate, released with thymidine, treated with colcemid (Invitrogen), swelled in hypo-osmolar potassium chloride, fixed, and stained with Sytox Green (MBL International) or DAPI (Vectashield). Single whole mouse chromosome paint probe 15 (Applied Spectral Imaging) was applied following the manufacturer’s recommended protocol.
μCT.
Mice were killed at 9 or 10 weeks of age. No contrast was used. A General Electric Medical Systems EVS-RS9 μCT scanned the lower half of the body. Planar reconstructions and three-dimensional rendering were achieved with Microview software (General Electric).
Tissue Analysis.
Knees and rib cages were harvested, fixed in 10% formalin in PBS, and decalcified in 5% formic acid for histology or fixed in 4% paraformaldehyde in PBS and decalcified in 25% EDTA (pH 7.4) at 37°C for histology, in situ hybridization, immunohistochemistry, β-gal staining, and LCM. Five- to 10-μm sections from paraffin-embedded tissue were stained with H&E or safranin-O. Six-micrometer sections were processed for radioactive in situ hybridization using [P33]-UTP-labeled antisense riboprobes as described (45). Probes for in situ hybridization were as follows: Col2 (46), ColX (47), Ihh (48), Ptch1 (49), and Ucma (50). β-Gal staining was performed on frozen sections as previously described (51) and counterstained with nuclear fast red.
For immunohistochemistry, 6-μm paraffin sections were pretreated with 10,000 units/mL hyaluronidase I (Sigma) for 30 min at 37°C, quenched with 3% H2O2 in PBS for 30 min, blocked in 10% goat serum in PBS for 30 min, and incubated overnight at 4°C with anti-10E4 antibody (Seikagaku), diluted 1:100 in blocking solution. Goat biotinylated secondary antibody against mouse IgM (Vector Laboratories) at a dilution of 1:100 was applied for 30 min. Detection was performed with Streptavidin-HRP (Perkin-Elmer) diluted 1:100 in PBS for 30 min followed by incubation with DAB for 5 min and then counterstaining with Methyl Green (Sigma).
LCM.
Six-micrometer paraffin sections were transferred to PALM MembraneSlides, dried, deparaffinized, and stained with Mayer’s hematoxylin and eosin. Clones of 1–20 cells were then captured with the PALM Microbeam System (Carl Zeiss MicroImaging) and digested in Proteinase K. PCR was performed directly on the heat-inactivated cell lysate with outer primers. Outer primer PCR products were then used as templates for subsequent nested PCRs using inner primers.
Supplementary Material
Acknowledgments
We thank Henry M. Kronenberg (Massachusetts General Hospital) for providing the Osx-CreERT mouse, Oliver Smithies (University of North Carolina at Chapel Hill) for pOSDUPDEL, and Matt F. Hockin (University of Utah) for the TATCre protein. This work was supported by the Orthopedic Research and Education Foundation 2003 Centerpulse Resident Grant for Growth Factor Research; by the Howard Hughes Medical Institute; by National Institutes of Health Grants EY-11298, EY-17168, and 5R37HD030701-16; and by Grant VO 620/9-1 from the Deutsche Forschungsgemeinschaft (to A.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1813.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910875107/DCSupplemental.
References
- 1.Wicklund CL, Pauli RM, Johnston D, Hecht JT. Natural history study of hereditary multiple exostoses. Am J Med Genet. 1995;55:43–46. doi: 10.1002/ajmg.1320550113. [DOI] [PubMed] [Google Scholar]
- 2.Bovée JV. Multiple osteochondromas. Orphanet J Rare Dis. 2008;3:3. doi: 10.1186/1750-1172-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J, et al. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- 4.Ligon AH, Potocki L, Shaffer LG, Stickens D, Evans GA. Gene for multiple exostoses (EXT2) maps to 11(p11.2p12) and is deleted in patients with a contiguous gene syndrome. Am J Med Genet. 1998;75:538–540. doi: 10.1002/(sici)1096-8628(19980217)75:5<538::aid-ajmg17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Wuyts W, et al. Mutations in the EXT1 and EXT2 genes in hereditary multiple exostoses. Am J Hum Genet. 1998;62:346–354. doi: 10.1086/301726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- 7.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97:668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senay C, et al. The EXT1/EXT2 tumor suppressors: Catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 2000;1:282–286. doi: 10.1093/embo-reports/kvd045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornemann DJ, Duncan JE, Staatz W, Selleck S, Warrior R. Abrogation of heparan sulfate synthesis in Drosophila disrupts the Wingless, Hedgehog and Decapentaplegic signaling pathways. Development. 2004;131:1927–1938. doi: 10.1242/dev.01061. [DOI] [PubMed] [Google Scholar]
- 10.Porter DE, Simpson AH. The neoplastic pathogenesis of solitary and multiple osteochondromas. J Pathol. 1999;188:119–125. doi: 10.1002/(SICI)1096-9896(199906)188:2<119::AID-PATH321>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.Knudson AG., Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard MA, et al. Diminished levels of the putative tumor suppressor proteins EXT1 and EXT2 in exostosis chondrocytes. Cell Motil Cytoskeleton. 2001;48:149–162. doi: 10.1002/1097-0169(200102)48:2<149::AID-CM1005>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Bovée JV, et al. EXT-mutation analysis and loss of heterozygosity in sporadic and hereditary osteochondromas and secondary chondrosarcomas. Am J Hum Genet. 1999;65:689–698. doi: 10.1086/302532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht JT, et al. Hereditary multiple exostosis and chondrosarcoma: Linkage to chromosome II and loss of heterozygosity for EXT-linked markers on chromosomes II and 8. Am J Hum Genet. 1995;56:1125–1131. [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht JT, et al. Hereditary multiple exostoses (EXT): Mutational studies of familial EXT1 cases and EXT-associated malignancies. Am J Hum Genet. 1997;60:80–86. [PMC free article] [PubMed] [Google Scholar]
- 16.Raskind WH, et al. Evaluation of locus heterogeneity and EXT1 mutations in 34 families with hereditary multiple exostoses. Hum Mutat. 1998;11:231–239. doi: 10.1002/(SICI)1098-1004(1998)11:3<231::AID-HUMU8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, et al. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- 18.Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132:5055–5068. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung PK, et al. Etiological point mutations in the hereditary multiple exostoses gene EXT1: A functional analysis of heparan sulfate polymerase activity. Am J Hum Genet. 2001;69:55–66. doi: 10.1086/321278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet. 2001;2:780–790. doi: 10.1038/35093564. [DOI] [PubMed] [Google Scholar]
- 21.Grégoire D, Kmita M. Recombination between inverted loxP sites is cytotoxic for proliferating cells and provides a simple tool for conditional cell ablation. Proc Natl Acad Sci USA. 2008;105:14492–14496. doi: 10.1073/pnas.0807484105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewandoski M, Martin GR. Cre-mediated chromosome loss in mice. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 23.Matsumura H, et al. _targeted chromosome elimination from ES-somatic hybrid cells. Nat Methods. 2007;4:23–25. doi: 10.1038/nmeth973. [DOI] [PubMed] [Google Scholar]
- 24.Lam KP, Rajewsky K. Rapid elimination of mature autoreactive B cells demonstrated by Cre-induced change in B cell antigen receptor specificity in vivo. Proc Natl Acad Sci USA. 1998;95:13171–13175. doi: 10.1073/pnas.95.22.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- 26.Joshi SK, Hashimoto K, Koni PA. Induced DNA recombination by Cre recombinase protein transduction. Genesis. 2002;33:48–54. doi: 10.1002/gene.10089. [DOI] [PubMed] [Google Scholar]
- 27.Grover J, Roughley PJ. Generation of a transgenic mouse in which Cre recombinase is expressed under control of the type II collagen promoter and doxycycline administration. Matrix Biol. 2006;25:158–165. doi: 10.1016/j.matbio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Logan M, et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 29.Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 31.Maes C, Kobayashi T, Kronenberg HM. A novel transgenic mouse model to study the osteoblast lineage in vivo. Ann N Y Acad Sci. 2007;1116:149–164. doi: 10.1196/annals.1402.060. [DOI] [PubMed] [Google Scholar]
- 32.Becker I, Becker KF, Röhrl MH, Höfler H. Laser-assisted preparation of single cells from stained histological slides for gene analysis. Histochem Cell Biol. 1997;108:447–451. doi: 10.1007/s004180050185. [DOI] [PubMed] [Google Scholar]
- 33.Ohno T, et al. Clonality in nodular lymphocyte-predominant Hodgkin’s disease. N Engl J Med. 1997;337:459–465. doi: 10.1056/NEJM199708143370704. [DOI] [PubMed] [Google Scholar]
- 34.Harvey M, et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 35.Lu W, et al. Late onset of renal and hepatic cysts in Pkd1-_targeted heterozygotes. Nat Genet. 1999;21:160–161. doi: 10.1038/5944. [DOI] [PubMed] [Google Scholar]
- 36.Hameetman L, et al. The role of EXT1 in nonhereditary osteochondroma: Identification of homozygous deletions. J Natl Cancer Inst. 2007;99:396–406. doi: 10.1093/jnci/djk067. [DOI] [PubMed] [Google Scholar]
- 37.Clément A, et al. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet. 2008;4:e1000136. doi: 10.1371/journal.pgen.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hameetman L, et al. Decreased EXT expression and intracellular accumulation of heparan sulphate proteoglycan in osteochondromas and peripheral chondrosarcomas. J Pathol. 2007;211:399–409. doi: 10.1002/path.2127. [DOI] [PubMed] [Google Scholar]
- 39.Hecht JT, et al. Heparan sulfate abnormalities in exostosis growth plates. Bone. 2002;31:199–204. doi: 10.1016/s8756-3282(02)00796-2. [DOI] [PubMed] [Google Scholar]
- 40.Delgado E, Rodríguez JI, Rodríguez JL, Miralles C, Paniagua R. Osteochondroma induced by reflection of the perichondrial ring in young rat radii. Calcif Tissue Int. 1987;40:85–90. doi: 10.1007/BF02555710. [DOI] [PubMed] [Google Scholar]
- 41.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6:801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Lin X, Gan L, Klein WH, Wells D. Expression and functional analysis of mouse EXT1, a homolog of the human multiple exostoses type 1 gene. Biochem Biophys Res Commun. 1998;248:738–743. doi: 10.1006/bbrc.1998.9050. [DOI] [PubMed] [Google Scholar]
- 43.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 44.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 45.Vortkamp A, et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 46.Kohno K, Martin GR, Yamada Y. Isolation and characterization of a cDNA clone for the amino-terminal portion of the pro-alpha 1(II) chain of cartilage collagen. J Biol Chem. 1984;259:13668–13673. [PubMed] [Google Scholar]
- 47.Jacenko O, LuValle P, Solum K, Olsen BR. A dominant negative mutation in the alpha 1 (X) collagen gene produces spondylometaphyseal defects in mice. Prog Clin Biol Res. 1993;383B:427–436. [PubMed] [Google Scholar]
- 48.Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 49.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 50.Tagariello A, et al. Ucma—A novel secreted factor represents a highly specific marker for distal chondrocytes. Matrix Biol. 2008;27:3–11. doi: 10.1016/j.matbio.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Lobe CG, et al. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.