Abstract
Schizophrenia is a major debilitating psychiatric disorder affecting approximately one percent of the population worldwide. A tremendous amount of effort has been expended in the past two decades to identify genes influencing susceptibility to this disorder. Although there is a strong trend towards integrating the data from various genetic studies and their related biological information into a comprehensive resource for many complex diseases, we have been unable to find such an effort for schizophrenia or any other psychiatric disorder yet. Here, we present Schizophrenia Gene Resource (SZGR), a comprehensive database with user-friendly web interface. SZGR deposits genetic data from all available sources including association studies, linkage scans, gene expression, literature, Gene Ontology (GO) annotations, gene networks, cellular and regulatory pathways, and microRNAs and their _target sites. Moreover, SZGR provides online tools for data browse and search, data integration, custom gene ranking, and graphical presentation. This system can be easily applied to other complex diseases, especially other psychiatric disorders. The SZGR database is available at http://bioinfo.vipbg.vcu.edu:8080/SZGR/.
Keywords: Schizophrenia, candidate gene, gene ranking, data integration, gene network, linkage, association, gene expression
Introduction
Schizophrenia is a major debilitating psychiatric disorder affecting approximately one percent of the population worldwide.1 It is commonly considered to be a complex disorder with multiple genetic and environmental factors involved; however, genetic factors impact substantially upon risk for developing the disease, with heritability estimates ~80%.2 The genetic approaches used so far to identifying risk genes or markers for schizophrenia have been largely inconclusive, as investigators often frustratingly found a low replication rate of significant markers or genes in the linkage or association studies, or found no clear connection between the risk to schizophrenia and structural changes in these susceptibility genes. It is likely that a number of genes, each of which contributes a small risk, interact with each other or with environmental risk factors to cause this psychiatric phenotype.3 Thus, collection and systematic annotations of candidate genes with genetic evidence from multiple studies is urgently needed for the examination of gene × gene (G×G) and gene × environment (G×E) interactions.
We have seen during the past two decades an exponential growth of vast amounts of biological data in schizophrenia genetics, including those generated by traditional positional cloning approach,4 individual gene/marker association studies and emerging genome-wide association studies,5–8 more than 32 genome-wide linkage scans and several meta-analyses,9, 10 and a large number of microarray experiments.11 Besides these genetic datasets, abundant biological information for the schizophrenia candidate genes can be extracted from public databases such as Gene Ontology annotations,12 protein-protein interaction (PPI) networks, and regulatory and cellular pathways.13 At present, there is a strong trend towards integrating the data from various genetic studies and their related biological information in the cellular systems so that promising candidate genes can be prioritized for follow up bioinformatics analysis and experimental verification. Some examples are National Cancer Institute (NCI) Cancer Gene Data Curation Project and a number of databases for specific categories of cancer (e.g. Tumor Suppressor Gene Database and Breast Cancer Database). For schizophrenia and the related psychiatric disorders, the VSD database focuses on variation data for publicly available schizophrenia candidate genes.14 This database seems no longer available, as its web link is not functional. Most recently, there is a SchizophreniaGene database that is specifically for the published association studies for schizophrenia.5 Another database, Sullivan Lab Evidence Project (SLEP), has been recently developed for the linkage and association evidence of genes or loci based on curation of the data.4 Each of these three databases focuses on specific genetic information for schizophrenia with only few computational tools available for the user. So far, we have been unable to find a comprehensive and integrative resource for schizophrenia.
Here, we present Schizophrenia Gene Resource (SZGR), a comprehensive database with user-friendly web interface. SZGR deposits genetic data collected from all the available sources including association studies, linkage scans, gene expression, literature, Gene Ontology (GO) annotations, gene networks, cellular and regulatory pathways, and microRNAs (miRNAs) and their _target sites. Besides, SZGR provides online tools for data integration and custom gene ranking, powerful data browse and search function, and graphical presentation. It has dynamic links to many public databases such as NCBI and the SchizophreniaGene. SZGR has been applied in several projects including schizophrenia gene network analysis and a large-scale genotyping project based on the prioritized candidate genes. This system can be easily applied to other complex diseases.
Database contents
One important feature of SZGR is its comprehensive collection of data from all major genetic studies for schizophrenia and systematic annotations. So far, we have collected data from seven major sources and categorized them into eight datasets. These datasets are association studies, three sets of meta-analysis of genome-wide linkage scans, meta-analysis of gene expression studies, high throughput literature search, genes by GO annotations, and genes by gene network features (Table 1). The data collection and curation was briefly described below. More details can be found on the SZGR web site and in our recent gene ranking study.15
Table 1.
Summary of datasets deposited in SZGR
| Gene set | Method | Number of genes | Reference |
|---|---|---|---|
| Association | Case-control association study | 281 | SchizophreniaGene Database: http://www.schizophreniaforum.org/ |
| GSMA_I | Meta-analysis of genome-wide linkage studies | 2158 | Lewis et al. (2003)9 |
| GSMA_IIA | Meta-analysis of genome-wide linkage studies | 2295 | Ng et al. (2009)10 |
| GSMA_IIE | Meta-analysis of genome-wide linkage studies | 1474 | Ng et al. (2009)10 |
| Expression | Meta-analysis of gene expression data sets | 726 | The SMRI Online Genomics Database: https://www.stanleygenomics.org/ |
| Literature | Literature search | 1682 | http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed |
| GO_Annotation | Mapping neuro- keywords | 1947 | Gene Ontology: http://www.geneontology.org/ |
| Gene_Network | Protein-protein interaction network | 1035 | HPRD,17 BIND,18 IntAct,19 MINT,20 Reactome21 and DIP22 |
| Core genes | Manual collection | 38 | Ross et al. (2006); Allen et al. (2008) |
| 75 genes by COR | Gene ranking | 75 | Sun et al. (2008)16 |
| 160 by Lewis et al. | Gene ranking | 160 | Sun et al. (2009)15 |
| 173 by Ng et al. | Gene ranking | 173 | Sun et al. (2009)15 |
GSMA_IIA and GSMA_IIE are meta-analysis of genome-wide linkage studies for all samples and European-ancestry samples only.
Association data
For association, we first collected the data from the recently established SchizophreniaGene database (http://www.schizophreniaforum.org/). The downloaded genotyping data was processed by a data cleaning and risk-allele evaluation pipeline developed in our recent combined odds ratio (OR) method.16 We selected the genes that had significant P value using our combined OR method or at least one positive association result in publication. Currently, there were 281 genes in this category, all of which have been genotyped with positive association signal in at least one study.
Linkage data
We selected linkage bins identified by the two genome scan meta-analyses (GSMA). The first GSMA was applied to data from 20 schizophrenia genome-wide linkage scans9 and identified 12 bins whose PAvgRnk and Pord are both <0.05. We obtained 2158 genes from these bins based on their genomic locations and gene annotations in NCBI (ftp://ftp.ncbi.nlm.nih.gov/gene/GeneRIF/) and defined them as “GSMA_I” in our database. The second GSMA was applied to 32 schizophrenia genome scans.10 We obtained genes from 10 bins whose PSR are <0.05 for all the samples and 6 bins only for European-ancestry samples. These two lists were defined as “GSMA_IIA” (2295 genes) and “GSMA_IIE” (1474 genes). The P value of each linkage bin was assigned to the genes within the bin.
Expression data
We downloaded gene expression data from the Stanley Medical Research Institute (SMRI, https://www.stanleygenomics.org/). The data is based on meta-analysis of 12 individual gene expression datasets from 988 microarrays for schizophrenia and bipolar disorder.11 We extracted 726 genes that were differentially expressed between schizophrenia post-mortem and control samples (P < 0.05) and considered them schizophrenia candidate genes.
Literature search
Co-occurrence of a gene and a schizophrenia-related keyword in an abstract may indicate that the gene is likely associated with schizophrenia. We performed a high throughput literature search based on this assumption using six schizophrenia-related keywords: “schizophrenia”, “schizophrenias”, “schizophrenic”, “schizophrenics”, “schizotypy” and “schizotypal”. We used the Linkout e-retrieval utility in NCBI Entrez Programming Utilities (http://eutils.ncbi.nlm.nih.gov/corehtml/query/static/eutils_help.html) to perform the literature search followed by manual checks of errors.
Gene Ontology
In psychiatric genetics as well as other complex disease studies, investigators have often selected functional candidates falling under the heading referred to as “the usual suspects” such as the genes suggested by neurotransmitter psychopharmacology. Searching genes by appropriate GO terms is useful and efficient for this purpose. A list of neuro-developmental terms was compiled based on expert recommendations (Supplementary Table S1). The related GO terms and their corresponding genes were identified based on keyword matching. We restricted those GO terms whose level were higher than 3 in GO tree because lower level GO terms tend to be non-specific (e.g. molecular function).
Protein-protein interaction network
Gene networks play important role in causing complex diseases. We first constructed the human interactome by integrating the experimentally verified PPI pairs from six databases: Human Protein Reference Database (HPRD),17 BIND,18 IntAct,19 MINT,20 Reactome21 and DIP.22 Then, we collected a small set of genes with “best” evidence so far to serve as “seeds” in network analysis. In this study, we selected 38 genes that had significant meta-analysis results5 or had been reviewed with extensive evidence.23 We named them core genes; the details are shown in SZGR. Among these 38 genes, 32 appeared in the human interactome. In network topology, proteins in the shortest path tend to have same or similar biological process.24, 25 For any pair of core genes, we identified its shortest path in the human interactome. Then, the genes in the shortest path were selected and scored (see next subsection). A total of 1035 genes were selected based on this network feature.
miRNA and _targets
Schizophrenia related miRNAs were collected from two independent studies which identified 18 miRNAs differently expressed in brain cortex of schizophrenia patients versus control samples.26, 27 We also collected and curated 87 non-schizophrenia specific brain expressed miRNAs from miRNA microarray expression studies and miRNA regulation surveys.26, 28, 29 Finally, we collected the miRNAs expressed in non-brain tissues from two large-scale miRNA expression atlas studies.30, 31 After removing schizophrenia or brain specific miRNAs, the remaining miRNAs were considered non-brain expressed.
The potential miRNA _target sites, family annotations, and sequence conservation information were extracted from the files downloaded from _targetScan (version 4.2, April 2008, http://www._targetscan.org/vert_42/). Then, the miRNA information was matched to schizophrenia candidate genes and made available in SZGR.
Summary
We collected experimental data from association, linkage and expression studies for schizophrenia, performed high throughput literature search and GO term analysis using the keywords or terms related to schizophrenia or neurodevelopment, and extracted schizophrenia candidate genes based on network features. Overall, the three datasets (“Association”, “Linkage”, and “Expression”) represent experimental data while the other three datasets (“Literature”, “GO_Annotation”, and “Gene_Network”) represent schizophrenia candidate genes with weak evidence. We also collected schizophrenia-specific or brain-specific miRNAs and their _target sites in the candidate genes. In total, there were 7855 non-redundant genes whose symbols could be found in the EntrezGene database (Table 1). The overlap between datasets is shown in Table 2.
Table 2.
Genes overlapped between datasets
| Number of genes overlapped |
||||||||
|---|---|---|---|---|---|---|---|---|
| Association | GSMA_I | GSMA_IIA | GSMA_IIE | Expression | Literature | GO_Annotation | Gene_Network | |
| Association | 281 | |||||||
| GSMA_I | 83 | 2158 | ||||||
| GSMA_IIA | 35 | 716 | 2295 | |||||
| GSMA_IIE | 22 | 301 | 733 | 1474 | ||||
| Expression | 15 | 85 | 58 | 34 | 1666 | |||
| Literature | 252 | 423 | 88 | 179 | 70 | 726 | ||
| GO_Annotation | 133 | 241 | 159 | 113 | 67 | 397 | 1947 | |
| Gene_Network | 71 | 130 | 76 | 57 | 40 | 244 | 238 | 1035 |
Database design and implementation
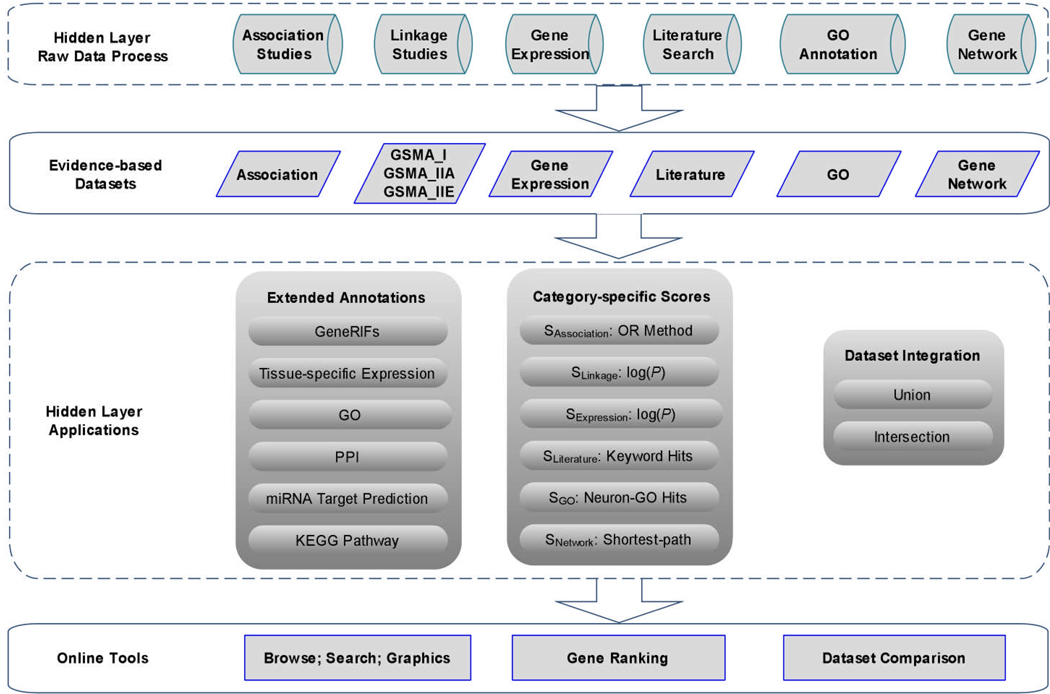
We designed the SZGR database using a multi-layer structure. As illustrated in Figure 1, the system includes two hidden layers for data process and computational tool development and two user-accessible platforms for data access and analysis. In the raw data process, we have done numerous data collection and curation, some of which was in a manual or semi-manual fashion. This includes data cleaning, cross-dataset mapping, data redundancy check, and reformatting. In the application layer, we developed tools for gene annotations (e.g. PPI network, KEGG pathway), gene ranking based on category-specific scoring algorithm, and dataset integration. These applications as well as the collected data are accessible to the end user.
Figure 1.
Flowchart of SZGR. It includes data collection, annotations, data integration, gene ranking and database development. GO: Gene Ontology. PPI: Protein-protein interaction. OR: odds ratio.
The multi-layer framework for SZGR makes it easy to modify settings or update data within each layer and to communicate between layers since each layer is independent. This is an important feature since many large-scale or genome-wide datasets are expected to be generated in the near future. This design allows us to add new datasets as well as to develop new computational tools easily. This system can be similarly applied to other complex diseases.
SZGR was implemented as a relational database using the open source MySQL database system and is freely accessible through a web interface developed in JSP technology. Each dataset is managed in MySQL database as a table that stores specific information while keys (e.g. dataset name, gene ID and PubMed ID) are extensively used for relational linking. The dynamic presentation of PPI networks was implemented by using the JAVA package provided by Medusa (http://coot.embl.de/medusa/). The gene ranking tool was implemented using JAVA language with the results being displayed graphically using JFree package (http://www.jfree.org/jfreechart/).
Web interface
We developed a user-friendly web interface for SZGR. The user may access all the data and perform analysis via the web interface (http://bioinfo.vipbg.vcu.edu:8080/SZGR/).
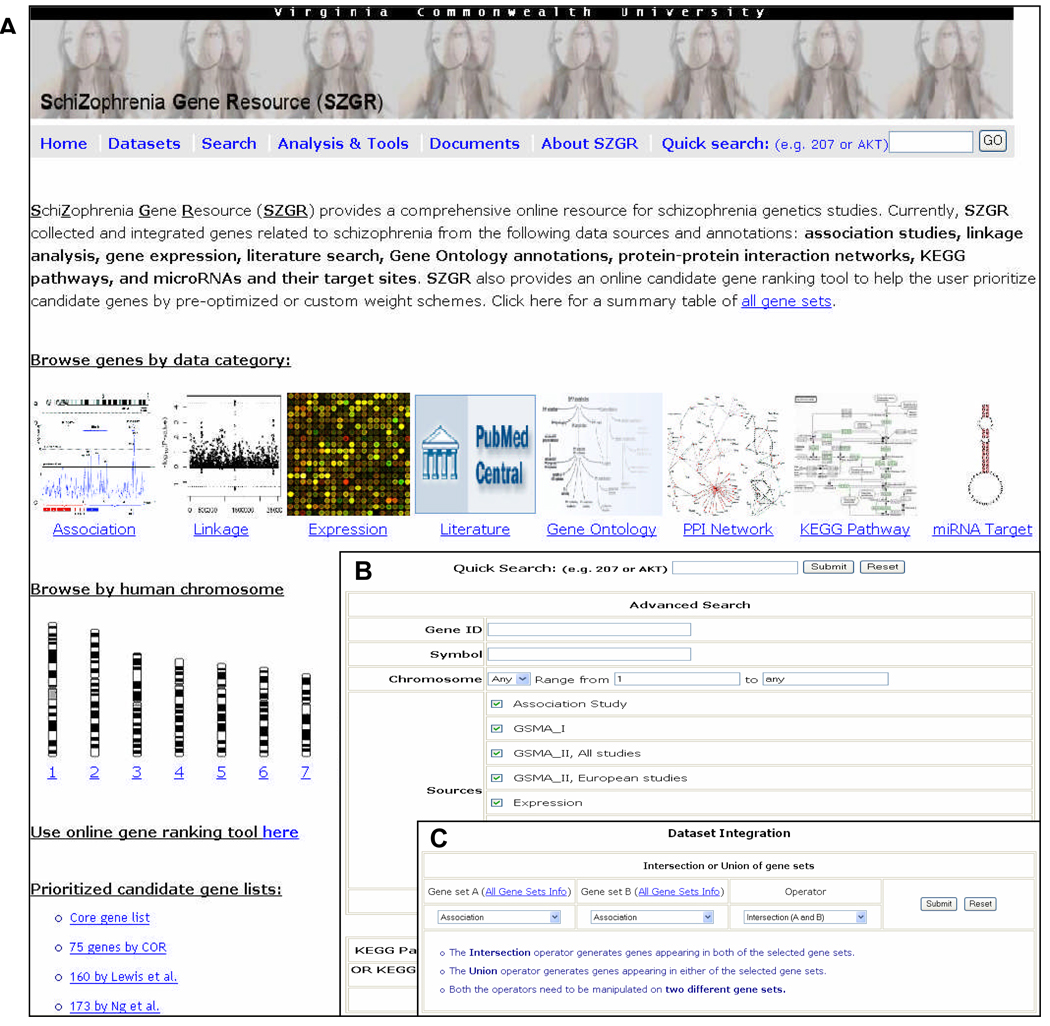
Data browse
In the main page (Figure 2A), the user may browse data by: (1) clicking one of the eight data categories (“Association”, “Linkage”, “Expression”, “Literature”, “GO_Annotation”, “Gene_Network”, “KEGG_Pathway”, and “miRNA__target”); (2) selecting one chromosome; (3) selecting one of the “Datasets” on the function bar on the top of the web page; or (4) clicking one of the four lists of prioritized candidate genes generated in our recent studies.
Figure 2.
SZGR web interface. (A) Home page. (B) Search page. (C) Data integration page.
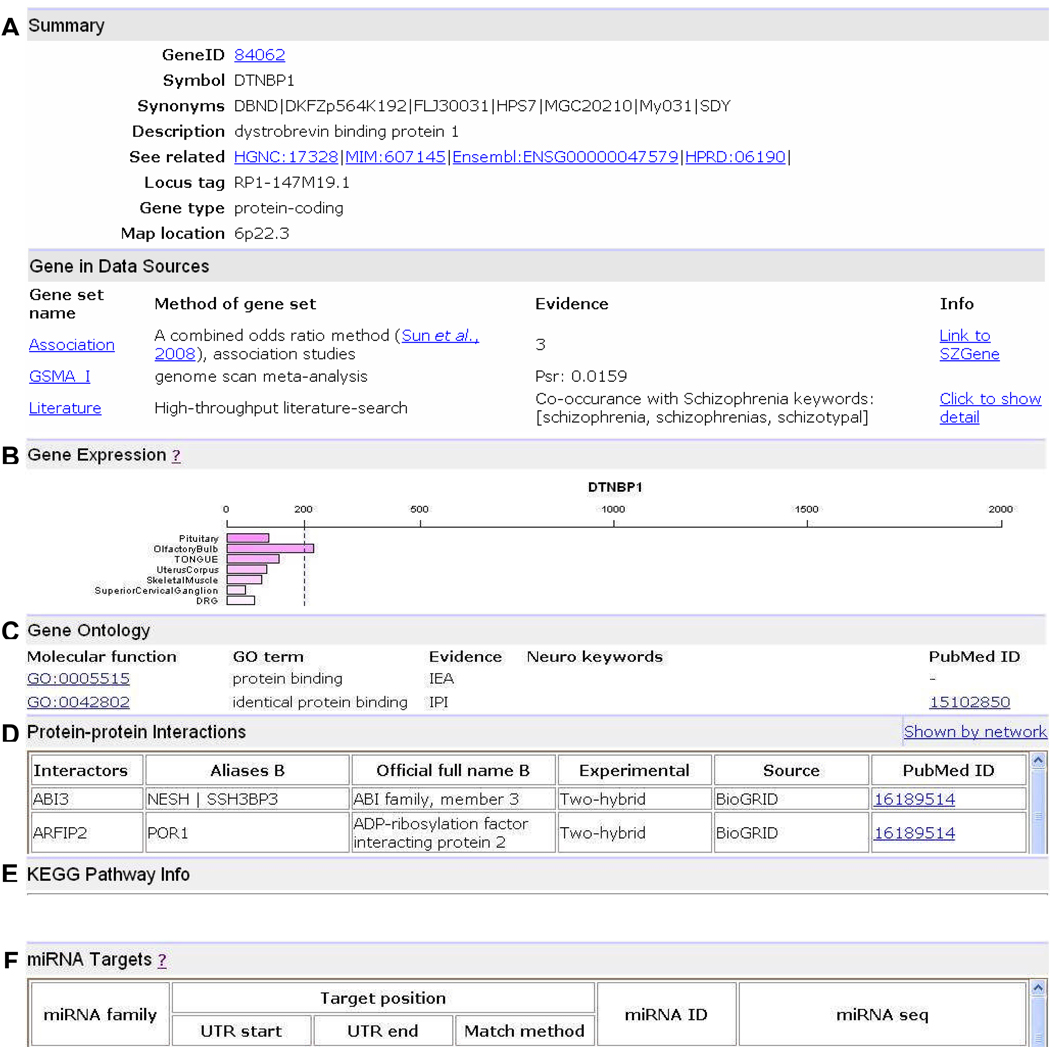
All datasets are relationally linked. Once the user clicks a gene ID, a detailed gene page is shown. It includes the following information.
(1) A summary for the gene, including gene symbol, synonyms, description, type, map location, and external links to other public databases (Figure 3A).
Figure 3.
An example gene page in SZGR. (A) Summary of the gene. (B) Data sources. (C) Gene expression profile. (D) Gene Ontology (GO) annotations. (E) KEGG pathway information. (F) miRNA _target sites. Due to the space limitation, only part of the content in each section is shown. Note that a gene (e.g. DTNBP1) may not have information in all categories.
(2) Data sources of the gene. This summarizes the evidence in six major data categories including a category-specific score and web link to the related databases when available (e.g. SchizophreniaGene database or PubMed) (Figure 3A).
(3) Gene expression profile in 79 human tissues extracted from the Gene Atlas (version 2).32 Gene expression in a tissue was measured using the arithmetic mean of the average difference (AD) values of their corresponding probe sets. The ADs for a gene are dynamically plotted in a single graph (Figure 3B).
(4) Gene Ontology annotations. The neuro-related GO terms are highlighted (Figure 3C).
(5) Protein-protein interactions between the protein encoded by the gene and other proteins in the human interactome. It presents local PPI environment by listing its direct interactors (distance 1) and distance-2 interactors (proteins that directly interact with the distance-1 proteins) (Figure 3D).
(6) KEGG pathways in which the gene involves (Figure 3E).
(7) miRNA _target sites. It includes the information such as miRNA families, prediction (e.g., the start and end positions of each predicted _target site) and an external link to the MiRBase database (http://microrna.sanger.ac.uk/http://microrna.sanger.ac.uk/) (Figure 3F).
Data search
SZGR provides multiple search options in a user-friendly environment. Besides a quick search function on the top right of the web page, the user may search by gene id, symbol, synonym, chromosomal region, specific data source, or by a user-defined combined setting (Figure 2B). It also provides an option to search genes involved in a pathway by pathway ID or name.
Data integration
Data integration includes two functions: union and intersection. Union is to combine the genes appearing in both gene sets, while intersection is to find genes common in both gene sets (Figure 2C). This simple tool may help the user quickly identify genes with specific evidence.
Gene ranking
Gene ranking algorithm
In SZGR, we extended and implemented a multi-dimensional evidence-based gene ranking algorithm developed in our recent study.15 In this algorithm, in each data category, each gene is assigned a category-specific score. There are four data categories (association, linkage, expression, and literature) in Sun et al.;15 here we extended to six data categories (GO and network). For most data categories, gene score is calculated by –log10 P when P values are available. For literature search, we assigned score for a gene based on the number of keywords being hit in the search. Similarly, we assigned a score for a gene based on the number of neuro-related GO terms annotated to the gene.
For gene network, the genes in the shortest path to a pair of core genes were assigned scores to measure their closeness to the phenotype (i.e. schizophrenia). We modified Wu et al.33 method to calculate the closeness of a gene in the shortest path to schizophrenia (i.e. core genes). The closeness of a gene g in the shortest path to a schizophrenia core gene is calculated by Gaussian kernel where g’ is the core gene and L is the distance between genes g and g’ in the shortest path. Therefore, the final score of gene g is the sum of its closeness to all core genes:
where C is the set of core genes.
Next, we searched an optimal weight matrix that weighs the score in each data category differently. The search of the optimal weight matrix was described in Sun et al.,15 which is based on two steps evaluated by the core genes and independent GWAS P values. The final score of a gene is calculated by
where i is the data category index and wi is the corresponding weight in the weight matrix.
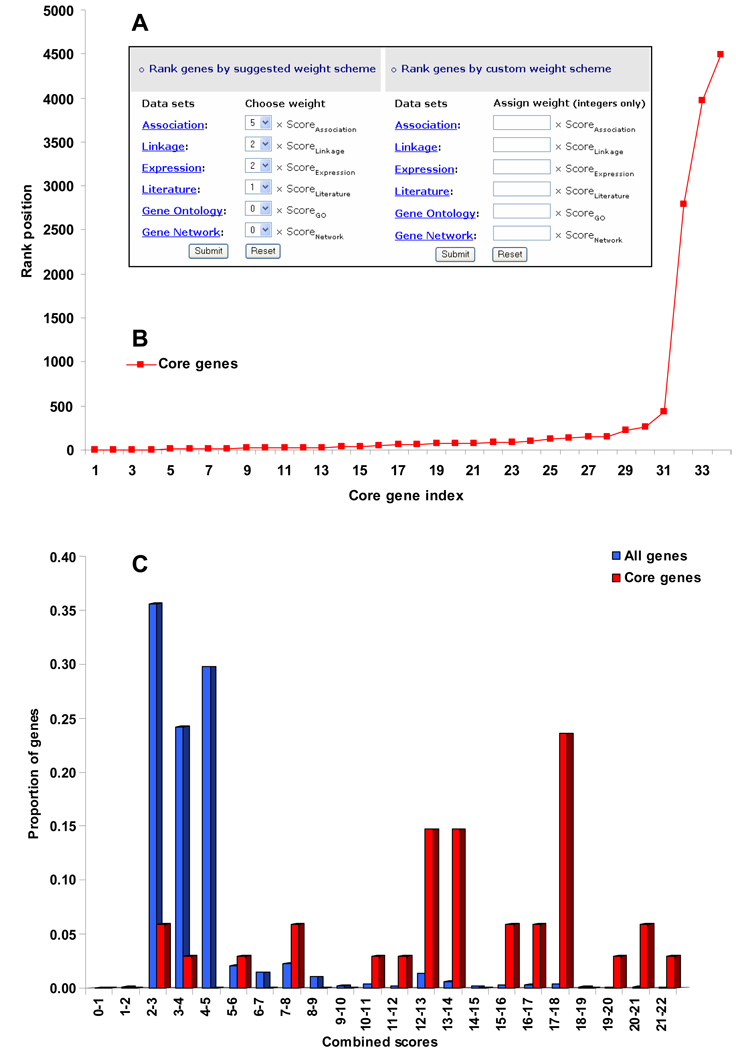
Gene ranking tool
SZGR provides online tool for gene ranking. Currently, it has two options (Figure 4A). The first one is based on the optimal weight scheme that was recommended in our recent multi-dimensional evidence-based candidate gene prioritization method.15 The score for each data category is initially calculated and then a combined score by weighing the category-specific scores is calculated. Genes are then ranked by their combined scores. The second option is custom weight scheme. The user may choose any weight for each data category based on his/her prior knowledge or special interest.
Figure 4.
Gene ranking and graphical presentation. (A) Online gene ranking tool. (B) Graphical presentation of rank positions of the core genes among all candidate genes. (C) Distribution of core and all genes by their scores.
To help the user evaluate and fine-tune the weight scheme, SZGR provides two graphical presentations of the ranking results. The first one is to show the rank positions of the core genes among all the ranked genes (Figure 4B). Since there has been no gold positive genes for schizophrenia yet (genes that have been confirmed to cause schizophrenia), we use core genes for this purpose, but they can be replaced by user-defined genes. Assuming that the core genes may have better evidence than other candidate genes, an efficient weight matrix is expected to rank the core genes, or their majority, on top of all candidate genes. The second one is a comparative distribution of the scores of the core genes and all genes. Genes are separated into different bins by their scores (e.g., 1–2, 2–3). An ideal distribution is that most of the core genes are ranked on the top while only few are ranked in the middle or at the bottom, as illustrated in Figure 4C.
Applications
Prioritized candidate genes
There are multiple approaches to select and prioritize candidate genes for complex diseases. A straightforward approach is to evaluate and weigh genetic significance information in multiple studies in one data category. We demonstrated this by ranking more than 500 genes in more than 2000 association studies and prioritizing 75 candidate genes using combined OR method.16 This gene list, named “75 genes by COR”, is available in SZGR.
We also demonstrated a multi-dimensional evidence-based gene prioritization approach for schizophrenia genetic data.15 By using the optimal weight matrix and evidence in four categories of data (association, linkage, expression, and literature), we prioritized 160 genes using the first version of GSMA results9 (named “160 by Lewis et al.”) and 173 genes using the just released second version GSMA results10 (named “173 by Ng et al.”). Both gene lists are available in SZGR.
Gene network analysis
The prioritized candidate genes can be further applied to follow up bioinformatics analysis and experimental verification. Here we demonstrated it by one example. For the 160 candidate genes, we extracted their network from the human interactome using the Steiner minimal tree algorithm.34 The network had 233 nodes including 135 prioritized genes (named SZGenes) and 98 new genes (named non-SZGenes). We examined association signal of these non-SZGenes in an independent genotyping project (unpublished data). In this project, a total of 3660 SNPs from 191 schizophrenia genes were genotyped in our Irish Study of High Density Schizophrenia Families (ISHDSF) sample. Among the 98 non-SZGenes, 6 genes were included for genotyping, three had P values <0.05. In one gene, five SNPs had P < 0.05 and the smallest P value was 0.00091.
Conclusion and future work
SZGR is a comprehensive resource for schizophrenia genetics. It includes all the major schizophrenia genetic datasets and their related biological annotations. We developed online tools for data access, gene ranking and bioinformatics analysis. To our understanding, this is the most comprehensive resource for schizophrenia, or such kind in psychiatric genetics.
The identification of potential schizophrenia susceptibility genes is expected to accelerate because of many genome-wide association studies (GWAS), digital gene expression profiling, epigenetics and epigenomics, and high-throughput proteomics. As in many other complex disorders and traits, we are following up the strong trend towards the integration of newly generated data and the use of this integrated data to generate lists of prioritized candidate genes. Besides the common disease/ common variant (CDCV) model, we are collecting and annotating the rare variants as well as structural variants (e.g., copy number variants) for schizophrenia, as such data may provide new insights on the molecular mechanisms.35, 36 Moreover, we will develop more computational tools for bioinformatics analysis such as gene network/pathway analysis. Finally, we are developing infrastructure allowing the user to deposit new datasets for user-driven data analysis and gene ranking. Our system design allows us to extend SZGR easily and flexibly.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Kenneth Kendler, Ayman Fanous, Brien Riley, and many other colleagues for their valuable discussions. This work was supported by grants from National Institute of Health, Thomas F. and Kate Miller Jeffress Memorial Trust Fund, and NARSAD Young Investigator Award to Z.Z.
Footnotes
Supplementary information
Supplementary information is available at the Molecular Psychiatry website (http://www.nature.com/mp).
References
- 1.Owen MJ, Craddock N, O'Donovan MC. Schizophrenia: genes at last? Trends Genet. 2005;21:518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 3.Burmeister M, McInnis MG, Zollner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- 4.Konneker T, Barnes T, Furberg H, Losh M, Bulik CM, Sullivan PF. A searchable database of genetic evidence for psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:671–675. doi: 10.1002/ajmg.b.30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams HJ, Owen MJ, O'Donovan MC. Schizophrenia genetics: new insights from new approaches. Br Med Bull. 2009 doi: 10.1093/bmb/ldp017. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009 doi: 10.1038/nature08185. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng MYM, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009 doi: 10.1038/mp.2008.135. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgs BW, Elashoff M, Richman S, Barci B. An online database for brain disease research. BMC Genomics. 2006;7:70. doi: 10.1186/1471-2164-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M, Zhuang YL, Xu Q, Li YD, Shen Y. VSD: a database for schizophrenia candidate genes focusing on variations. Hum Mutat. 2004;23:1–7. doi: 10.1002/humu.10289. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Jia P, Fanous AH, Webb BT, van den Oord EJCG, Chen X, et al. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases – schizophrenia as a case. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp428. Advance access published on July 14, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- 17.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW. BIND--The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 21.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Witten TM, Bonchev D. Predicting aging/longevity-related genes in the nematode Caenorhabditis elegans. Chem Biodivers. 2007;4:2639–2655. doi: 10.1002/cbdv.200790216. [DOI] [PubMed] [Google Scholar]
- 25.Managbanag JR, Witten TM, Bonchev D, Fox LA, Tsuchiya M, Kennedy BK, et al. Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity. PLoS ONE. 2008;3:e3802. doi: 10.1371/journal.pone.0003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 27.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11) Biochemistry (Mosc) 2007;72:578–582. doi: 10.1134/s0006297907050161. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R, Su B. MicroRNA regulation and the variability of human cortical gene expression. Nucleic Acids Res. 2008;36:4621–4628. doi: 10.1093/nar/gkn431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein P, Ravi RA. Nearly best-possible approximation algorithm for node-weighted Steiner trees. J Algorithms. 1995;19:104–115. [Google Scholar]
- 35.The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.