Abstract
Memory formation requires changes in gene expression, which are regulated by the activation of transcription factors and by changes in epigenetic factors. Poly[ADP]-ribosylation of nuclear proteins has been postulated as a chromatin modification involved in memory consolidation, although the mechanisms involved are not well characterized. Here we demonstrate that poly[ADP]-ribose polymerase 1 (PARP-1) activity and the poly[ADP]-ribosylation of proteins over a specific time course is required for the changes in synaptic plasticity related to memory stabilization in mice. At the molecular level, histone H1 poly[ADP]-ribosylation was evident in the hippocampus after the acquisition period, and it was selectively released in a PARP-1-dependent manner at the promoters of cAMP response element-binding protein and nuclear factor-κB dependent genes associated with learning and memory. These findings suggest that histone H1 poly[ADP]-ribosylation, and its loss at specific loci, is an epigenetic mechanism involved in the reprogramming of neuronal gene expression required for memory consolidation.
Introduction
During cognitive processes, changes in gene expression are required to store information (Kandel, 2001), and thus the regulation of transcription is critical for this process. Although many studies have focused on the activation of transcription factors in memory consolidation (Alberini, 2009), recent studies have demonstrated the role of chromatin remodeling in cognitive processes (Roth and Sweatt, 2009). In mammals, alterations to the core histone acetylation–deacetylation balance (Levenson et al., 2004a; Vecsey et al., 2007; Fontán-Lozano et al., 2008), in histone methylation (Gupta et al., 2010), and of DNA methylation (Miller and Sweatt, 2007; Lubin et al., 2008) have been demonstrated that can alter memory consolidation.
Poly[ADP]-ribosylation is a transient modification of many nuclear proteins, including histones, transcription factors, RNA polymerase II (RNA pol II), topoisomerases, and high-mobility group proteins, which regulates their binding to DNA (Schreiber et al., 2006). Poly[ADP-ribose] polymerase 1 (PARP-1) is activated by stressful stimuli that damage DNA (Virág and Szabó, 2002), although it can also be induced by other physiological stimuli. Depolarization and exposure of cortical neurons to neuronal growth factor have been shown to activate PARP-1 in the absence of DNA damage (Homburg et al., 2000; Visochek et al., 2005). Moreover, it was shown recently that PARP-1 activation is required for long-term neuronal plasticity in Aplysia and mice (Cohen-Armon et al., 2004; Goldberg et al., 2009; Hernández et al., 2009). However, the mechanisms by which PARP-1 operates in learning and memory are not well known. Our results show that changes in gene expression needed for consolidation of object recognition (OR) memory require PARP-1 activation in hippocampus. Moreover, training-dependent PARP-1 activation provokes histone H1 poly[ADP]-ribosylation and its release from specific gene promoters required for memory consolidation. These results indicate that poly[ADP]-ribosylation of histone H1 is another epigenetic modification contributing to memory coding at the genome level.
Materials and Methods
Animals.
Eight-week-old male Swiss mice weighing between 25 and 30 g were maintained on a 12 h light/dark cycle. Behavioral and electrophysiological studies were all performed in accordance with the European Union Council guidelines (86/609/EU) and Spanish regulations for the use of laboratory animals in chronic experiments (BOE 67/8509-12, 1988). Furthermore, all experiments were approved by the local institutional animal care committee.
Drug administration.
Thieno[2,3-c]isoquinolin-5-one (Tiq-A) (Sigma-Aldrich), a PARP-1 antagonist (0.5 mg/kg), or an equivalent volume of the vehicle alone was injected intraperitoneally. For local administration of the drug, the animals were anesthetized and secured in a stereotaxic frame, and guide cannulae were implanted bilaterally directed to the hippocampus as described previously (Fontán-Lozano et al., 2007). Two stainless-steel guide cannulae were implanted at the following coordinates for the hippocampus (relative to bregma): anteroposterior (AP), −2.2 mm; lateral (L), ±1.5 mm; and ventral (V), −1.1 mm. Between infusions, the cannulae were closed with a dummy cannulae and the mice were allowed to recover for at least 10 d before habituation to the testing arena began. Five minutes before performing the behavioral tests, 0.5 μl of a 250 nm solution of Tiq-A was microinjected over 60 s through each cannula, and the infusion cannulae were left for an additional 60 s to minimize backflow.
Behavioral assays.
Mice were tested in a rectangular arena (55 × 40 × 40 cm) located in a room with dim lighting and constant background noise. In the object recognition protocol, two different objects were placed in the arena during the training phase. After a delay of 1 or 24 h, one object was changed for a novel object. The aim was to test the animal's memory of the original objects by measuring the amount of time spent exploring the novel object versus the familiar one. The objects selected were plastic pieces with different forms, and they were thoroughly cleansed between trials to ensure the absence of olfactory cues. Before the experiment, mice were habituated to the arena in the absence of objects for 20 min each day over 2 d, and, on the day of testing, mice were allotted 15 min to explore the two objects. Retention tests were performed either 1 or 24 h later by placing the mice back in the arena for 10 min and by randomly exchanging one of the familiar objects with a novel one. The time spent exploring each object was recorded, and the relative exploration of the novel object was expressed as the discrimination index [D.I. = (tnovel − tfamiliar)/(tnovel + tfamiliar)]. The criteria for exploration were based strictly on active exploration, in which the mouse had both forelimbs within a 1.5 cm circle around the object, with its head oriented toward it, or when the animal touched the object with its vibrissae.
For RNA and protein analyses, mice were habituated to the arena in the absence of objects for 20 min/d on 2 consecutive days. On the day of testing, untrained and trained mice were left for 15 min to explore the arena with or without objects, after which the animals were killed at the times indicated.
Electrophysiology.
Animals were anesthetized with 0.8–3% halothane delivered through a home-made mask (AstraZeneca). Halothane was administered from a calibrated Fluotec 5 (Fluotec-Ohmeda) vaporizer at a flow rate of 1–4 L/min oxygen. Once anesthetized, the animals were implanted with electrodes made of 50 μm Teflon-coated tungsten wires (Advent Research Materials Ltd.). Using stereotaxic coordinates (Paxinos and Franklin, 2001), bipolar stimulating electrodes were implanted into the Schaffer collateral–commissural pathway of the dorsal hippocampus (from bregma: AP, 1.5 mm; L, 2.2 mm; depth from brain surface: 1.0–1.5 mm). Two recording electrodes were also implanted in the ipsilateral stratum radiatum of the CA1 area (from bregma: AP, 2.2 mm; L, 2.2 mm; depth from brain surface: 1.0–1.5 mm). The final position of the hippocampal stimulating and recording electrodes was determined by online recording procedures (see below). The four wires were connected to a four-pin socket (RS-Amidata), whereas the ground wire was connected to the recording system with a single wire. A 0.1 mm bare silver wire was fixed to the skull as a ground. Sockets were fixed to the skull with the help of two small screws and dental cement (for more details, see Gruart et al., 2006; Fontán-Lozano et al., 2007). Recordings were performed using Grass P511 differential amplifiers with a bandwidth of 0.1 Hz to 10 kHz (Grass-Telefactor).
All in vivo recordings were performed at least 7 d after implantation of the electrodes to allow the animals to recover from surgery. To evoke long-term potentiation (LTP), each animal received five pulse trains (200 Hz, 100 ms) at a rate of 1/s, a protocol that was administered either six times at 1 min intervals. The hippocampal activity recorded was stored digitally on a computer through an analogical-to-digital converter (CED 1401 Plus; Cambridge Electronic Design) at a sampling frequency of 11–22 kHz and with an amplitude resolution of 12 bits. The Spike 2 and SIGAVG software (Cambridge Electronic Design) were adapted to represent field EPSP (fEPSP) recordings. The slope of the evoked fEPSPs was collected as the first derivative (volts per second) of the fEPSP records (volts). To calculate this derivative, five successive evoked field synaptic potentials at intervals of 5 min were averaged, and the mean value of the slope was determined for the rise-time period (i.e., the period of the rise between the initial 10% and the final 10% of the evoked field potential). For paired-pulse facilitation (PPF), two stimuli of an intensity that evoked 35–40% of the maximum response fEPSP were delivered with an interstimulus interval of 20–200 ms. The percentage of facilitation was calculated as (slope S2/slope S1) × 100.
Two-dimensional gel electrophoresis.
Hippocampi were lysed in cell lysis buffer (7 m urea, 2 m ThioUrea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 40 mm PlusOne Tris, and 60 mm DTT), and, after centrifugation, the proteins were precipitated with 10% TCA and 0.2% DTT. The resulting pellet was washed twice with 1 ml of chilled acetone and dried for 15 min at room temperature. Subsequently, the proteins were resuspended in a rehydration solution [7 m ThioUrea, 2 m CHAPS, 4% DTT, and 0.8% immobilized pH gradient (IPG) buffer] and quantified using an RC-DC Protein Assay (Bio-Rad) according to the instructions of the manufacturer. One-dimensional electrophoresis was performed on an Inmobiline Drystrip pH 3-11 NL (GE Healthcare) using 80 μg of protein per strip. The strips were focused in an IPGphor system for 22 h, and then two-dimensional PAGE was performed on 10% duracryl acrylamide gels (Genomics Solutions) run in an Ettan Dalt VI system (GE Healthcare). After two-dimensional PAGE resolution, the proteins were analyzed in Western blots probed with an anti-histone H1 antibody (1:1000 dilution; Santa Cruz Biotechnology).
Immunoprecipitation and Western blotting.
Hippocampal protein extracts were immunoprecipitated using an immunoprecipitation kit (Roche) according to the instructions of the manufacturer. Western blots were probed with antibodies against poly[ADP]-ribose (PAR) polymer (Biomol) and histone H1 (Santa Cruz Biotechnology) diluted 1:1000. As an internal control for immunoprecipitation, a 10% sample of the protein used for immunoprecipitation was assayed for PAR polymer, histone H1, and actin immunoreactivity (Santa Cruz Biotechnology).
Immunohistochemistry and histological analysis.
For immunohistochemistry (IHQ), antibodies against PAR (Biomol) and histone H1 (Santa Cruz Biotechnology) were used at dilutions of 1:1000 and 1:3000, respectively. Antibody staining was visualized with H2O2 and diaminobenzidine, and the tissue sections were examined by dark-field microscopy. To minimize variability, at least five sections per animal were analyzed on a bright-field DMRB RFY HC microscope (Leica). For IHQ quantification, sections corresponding to stereotaxic coordinates 21.34 to 22.80 mm with respect to bregma were analyzed.
Chromatin immunoprecipitation.
The chromatin immunoprecipitation (ChIP) assay was performed as described by Guan et al. (2002), with some modifications. The hippocampus was dissected out, fixed with 4% paraformaldehyde for 2 h, and washed in 0.125 m glycine and PBS, for 10 min each at 4°C, before it was homogenized and sonicated to obtain DNA fragments of 600 bp (average size). Specific antibodies, against histone H1 (Santa Cruz Biotechnology) or RNA polymerase II (Santa Cruz Biotechnology), were used for chromatin immunoprecipitation, and the presence of specific gene promoters was analyzed by semiquantitative PCR using the primers listed in the supplemental data (available at www.jneurosci.org as supplemental material). Input and immunoprecipitated DNA amplification reactions were run in triplicate in three independent experiments.
Reverse transcription-PCR analysis of mRNA.
Total RNA from brain tissue was extracted with the Tripure reagent (Roche Products) and a minimum of six animals per group, collected from at least two different experimental sessions, were used for each experiment. More detailed information on the primers can be found in the supplemental Experimental Procedures (available at www.jneurosci.org as supplemental material). Arbitrary units were calculated and compared with glyceraldehyde-3-phosphate dehydrogenase (gapdh) expression.
Statistical analyses.
The results were analyzed using the SPSS package for Windows. Unless otherwise stated, the data are represented as mean ± SEM values. Data were analyzed with one-way and two-way ANOVA, and the t test was used for post hoc comparisons.
Results
Object recognition training session increased protein poly[ADP]-ribosylation in the hippocampus and perirhinal cortex
The OR memory task is a one-trial model of learning and memory that requires new protein synthesis for memory consolidation (Romero-Granados et al., 2010). Object recognition memory is a phenomenon in which the perirhinal cortex is involved in object familiarization, whereas the hippocampus participates both in the formation of the relationship between objects and spatial cues and in the consolidation of the information acquired (Brown and Aggleton, 2001; Dere et al., 2007; Fontán-Lozano et al., 2007). To study the pattern of poly[ADP]-ribosylation of proteins in OR memory formation, we killed mice immediately after a 15 min training session and processed their brains for PAR polymer immunohistochemistry (Fig. 1). An increase in the PAR polymer was evident in the hippocampus (Fig. 1A) and perirhinal cortex (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) from the trained animals when compared with habituated mice [CA1: 13.31 ± 2.04 and 30.28 ± 1.33 arbitrary units (A.U.) for habituated and trained mice, respectively; F(8,1) = 9.832, p < 0.001; CA3: 26.93 ± 2.54 and 36.34 ± 2.01 A.U. for habituated and trained mice, respectively; F(8,1) = 8.42, p = 0.017; dentate gyrus: 24.06 ± 2.41 and 22.66 ± 2.81 A.U. for habituated and trained mice, respectively; F(8,1) = 0.14, p = 0.715; and perirhinal cortex: 3.85 ± 0.50 and 10.58 ± 0.65 cells per 100 μm2 for habituated and trained mice, respectively; F(8,1) = 66.394, p < 0.001]. In Western blots, there appeared to be 1.95 ± 0.16-fold more PAR polymer in the hippocampal protein extracts from trained mice compared with untrained mice (t(6) = 11.19, p < 0.01) (Fig. 1B). Furthermore, the formation of PAR polymer induced by training was blocked by Tiq-A (0.5 mg/kg), a PARP-1 inhibitor, when injected 15 min before the OR training session. These results suggest that PAR polymer formation induced by OR training requires PARP-1 activation.
Figure 1.
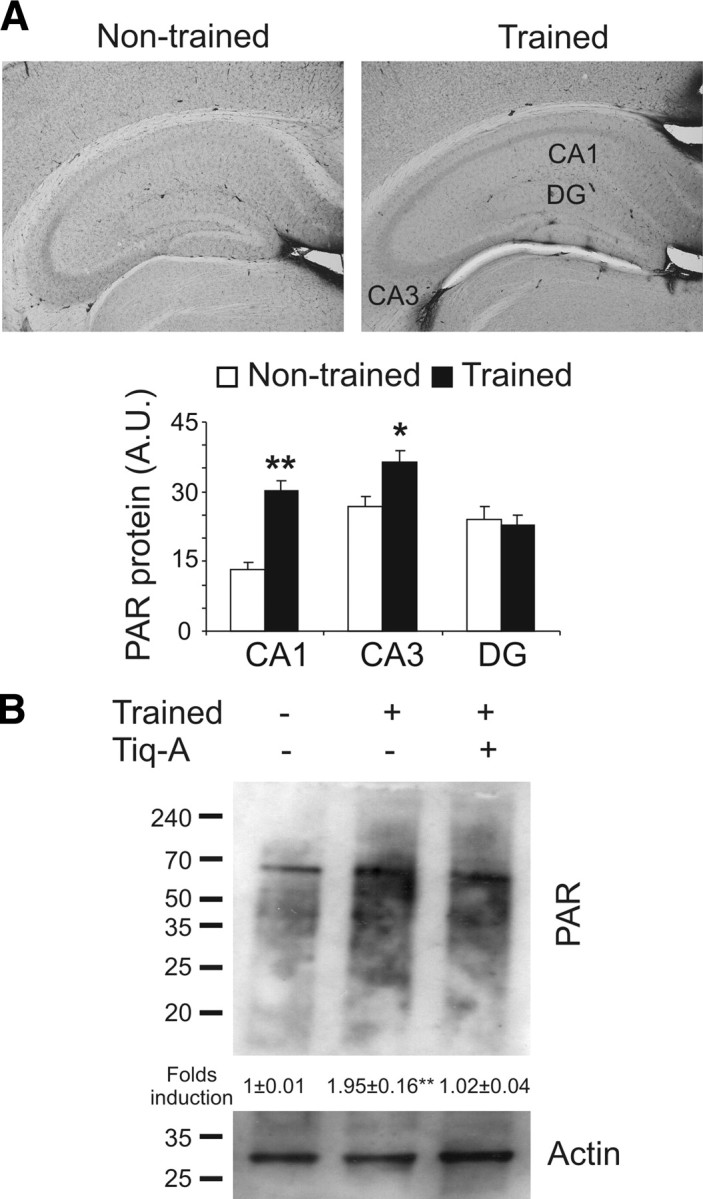
Synthesis of the PAR polymer is induced after object recognition training. A, The levels of PAR polymer were determined by immunohistochemistry in brains from mice killed 15 min after a 15 min OR training session (n = 5 mice per group). Bar chart illustrating the quantification of PAR polymer immunoreactivity in the CA1, CA3, and dentate gyrus (DG) of the hippocampus in habituated (white bars) and trained (black bars) mice. B, Representative Western blot probed for protein poly[ADP]-ribosylation in the hippocampus of mice administered the PARP-1 inhibitor Tiq-A or the vehicle alone and killed 15 min after a 15 min OR training session. Densitometry of three independent experiments is shown. Asterisks indicate significant differences in the densitometry between habituated and trained mice; *p < 0.05; **p < 0.01.
PARP-1 activation is required for OR consolidation
To evaluate the function of the PAR polymer in OR memory, we systemically administered Tiq-A 0.5 h before or 2 h after the training session ended (Fig. 2A). The pretraining injection of Tiq-A resulted in a long-term OR memory (LTM) deficits in tests performed 24 h after the OR training session (the D.I. values were 0.36 ± 0.05 and −0.01 ± 0.09 for the vehicle- and Tiq-A-injected mice, respectively), as suggested by the session × treatment interaction (F(42,5) = 14.03, p < 0.001). In contrast, Tiq-A did not have any significant effect on the short-term OR memory (STM) tested 1 h after the OR training session (D.I. values of 0.44 ± 0.05 and −0.44 ± 0.06 for vehicle- and Tiq-A injected mice, respectively). Interestingly, when Tiq-A was administered 2 h after the training session, no effects on STM or LTM were observed (Fig. 2B). Furthermore, no significant differences in exploratory activity were found between treatments (supplemental Table 1, available at www.jneurosci.org as supplemental material).
Figure 2.
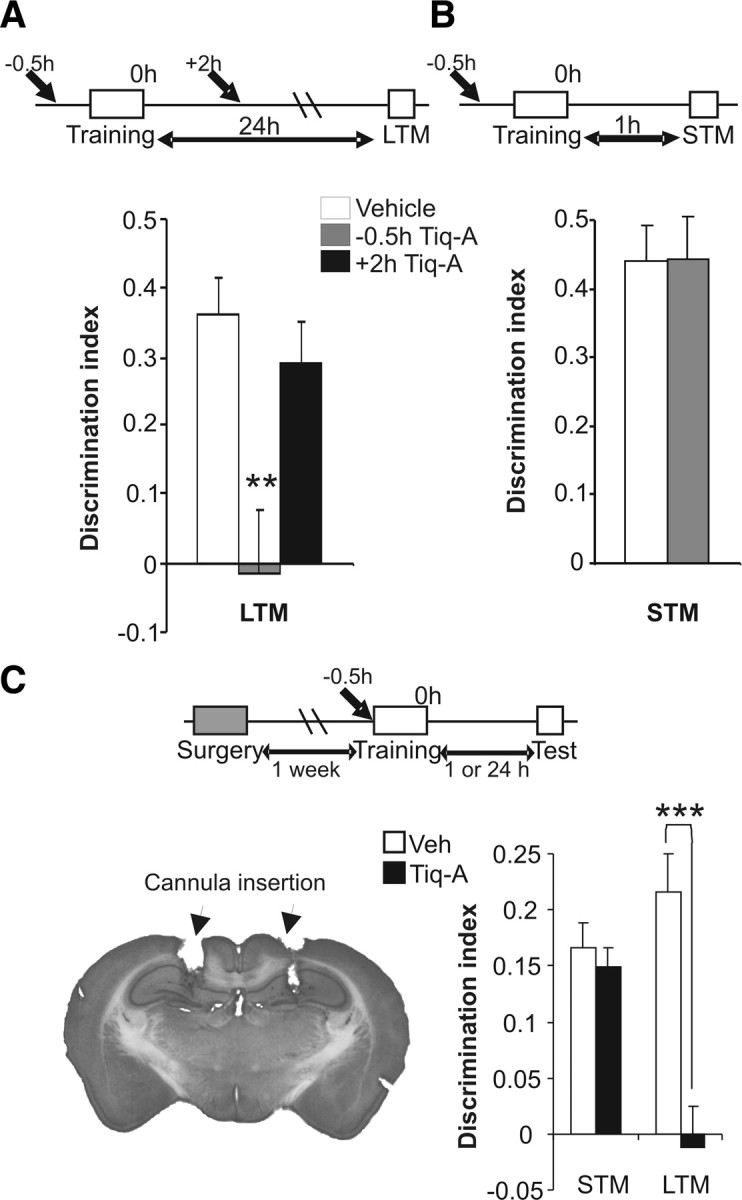
Hippocampal PARP-1 activation is required for object recognition memory consolidation. A, B, Mice were subjected to a 15 min OR training session, and 1 or 24 h after training, their STM and LTM, respectively, was tested. To test the role of the PAR polymer on memory consolidation, either 0.5 mg/kg Tiq-A or saline was injected at varying times from the end of the training session (from −0.5 to +2 h). The bar chart illustrates the discrimination indices calculated as (tnovel − tfamiliar)/(tnovel + tfamiliar) during LTM (A) and STM (B) sessions (n = 8 for each group). The bars in white represent saline administration, and gray and black bars represent the different times of Tiq-A administration. C, The effects of Tiq-A infusion directly into the hippocampus of mice. Tiq-A was administered 5 min before a 15 min training session, and the STM and LTM were assayed. Left, Representative microphotograph of the cannula located in the hippocampus. Right, The bar chart illustrates the discrimination indices during STM and LTM (n = 10 mice per group). **p < 0.01; ***p < 0.001.
To confirm the requirement of PARP-1 activation for OR consolidation in the hippocampus, we implanted bilateral cannula into this brain area. Tiq-A infusion through the cannula to the hippocampus of mice 5 min before the training protocol did not affect STM but it did disrupt LTM when compared with the values obtained after the infusion of saline (for hippocampus: t(18) = 4.53, p < 0.001) (Fig. 2C). Additionally, hippocampal administration of ADP–HDP (100 μm), an inhibitor of an enzyme that degrades PAR polymers (the poly[ADP]-ribose glycohydrolase), 5 min before the OR training session facilitated OR acquisition and consolidation (supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
All these data indicate that, during the training session and for up to 2 h after, PARP-1 activity is required for OR memory consolidation in the hippocampus.
Long-lasting changes in synaptic efficacy required PARP-1 activation
The hippocampus is a cortical region involved in information processing and memory consolidation (Squire and Zola-Morgan, 1991). Hippocampal electrophysiology in freely moving mice was studied by implanting stimulating electrodes into the Schaffer collaterals and a recording electrode in the apical dendrites of the CA1. To study the role of PARP-1 in glutamatergic basal synaptic transmission, PPF with interpulse intervals from 50 to 200 ms was performed in vehicle- and Tiq-A-injected mice. In vehicle-injected mice, the PPF was maximum at an interpulse interval of 50 ms, and it diminished to basal levels with a 200 ms interpulse interval (F(10,2) = 19.14, p < 0.001). Significantly, the administration of Tiq-A did not induce any changes in PPF at the interpulse intervals studied (Fig. 3A), suggesting that the CA3–CA1 synapse was not functionally affected by Tiq-A.
Figure 3.
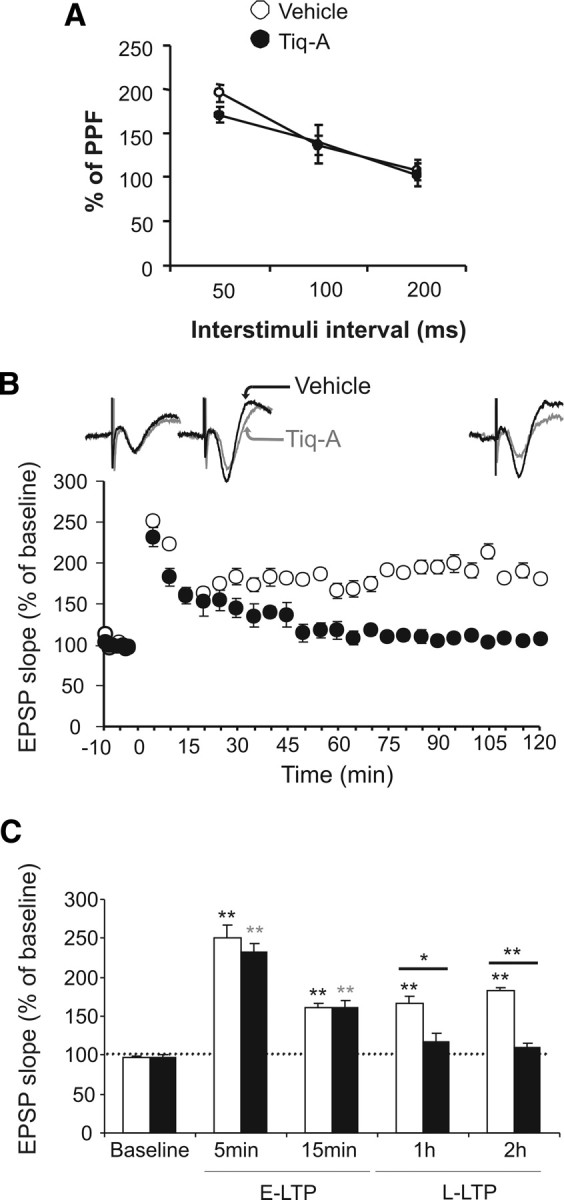
PARP-1 inhibition blocked long-lasting changes in synaptic efficacy. A, Basal excitatory neurotransmission was measured using paired-pulse facilitation with interpulse intervals from 50 to 200 ms in the presence or absence of Tiq-A. The percentage of paired-pulse facilitation as a function of the interpulse interval in mice receiving Tiq-A or the vehicle alone (white circle, vehicle; black circle, Tiq-A mice; n = 6 for each group) is presented. B, Changes in the slope of the fEPSP induced by six HFS [5 trains (200 Hz, 100 ms) of pulses at a rate of 1/s]. Note that the evoked long-lasting LTP lasted for up to 2 h in the vehicle-administered mice (white) but not in Tiq-A-administered mice (black; n = 6 for each group). Representative recordings at baseline and 15 or 120 min after HF from mice that received Tiq-A or the vehicle alone. C, Summary of the changes in fEPSP slope at different times after six HFS trains: baseline, 5 and 15 min after HFS [early LTP (E-LTP)], and 1 and 2 h after HFS [long-lasting LTP (L-LTP)]. All data are shown as mean values ± SEM. *p < 0.05 and **p < 0.01 indicates statistically significant difference between the vehicle- and Tiq-A-injected mice at different times after HFS.
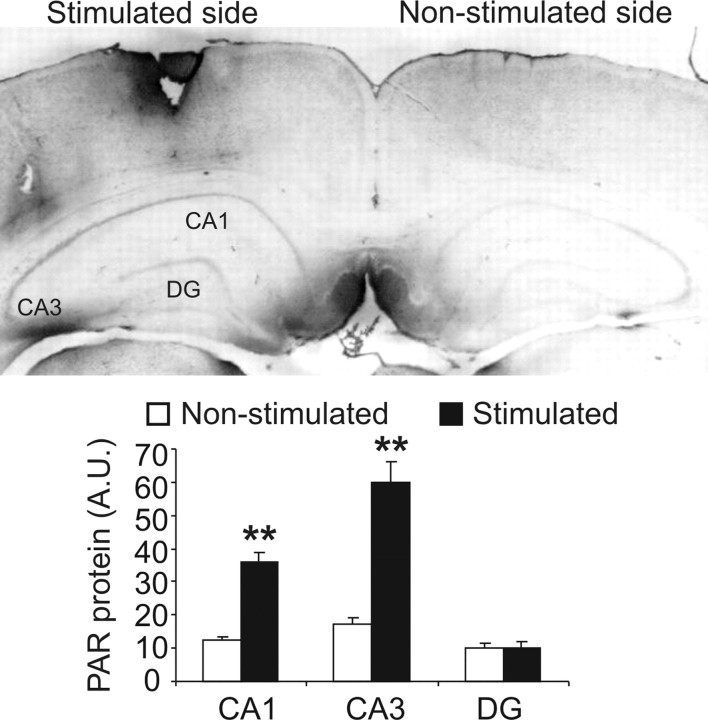
Because long-term changes in synaptic efficacy in the hippocampus seems to be necessary for learning and memory (Daoudal and Debanne, 2003; Gruart et al., 2006; Clarke et al., 2010), we examined the role of PARP-1 in the long-lasting changes of synaptic transmission efficacy induced by high-frequency stimulation (HFS) at the CA3–CA1 synapses. Tiq-A administration before six HFS protocols (HFS, consisting of five trains at 200 Hz, lasting 100 ms, and presented at a rate of 1/s) provoked a deficit in long-LTP when compared with vehicle-injected mice tested 1 or 2 h after application of HFS protocol as suggested by the time × treatment interaction (F(50,9) = 6.19, p < 0.001; the fEPSP slope 1 h after HFS was 165.95 ± 8.95 and 117.57 ± 10.09 for vehicle- and Tiq-A-injected mice, respectively, and 2 h after HFS was 182.22 ± 4.21 and 110.54 ± 5.21 for vehicle- and Tiq-A-injected mice, respectively) (Fig. 3B,C). In contrast, Tiq-A did not have a significant effect on the short-LTP tested 5 or 15 min after administration of the HFS protocol (the fEPSP slope 5 min after HFS was 251.16 ± 10.87 and 232.93 ± 11.66 for vehicle- and Tiq-A-injected mice, respectively, whereas 15 min after HFS, it was 161.04 ± 6.23 and 160.26 ± 10.02). Furthermore, entorhinal cortex stimulation that induced LTP in the hippocampus also augmented the PAR polymer in the hippocampal CA1 and CA3 fields. Indeed, the PAR polymer densitometry in the CA1 field augmented nearly threefold after stimulation (12.55 ± 0.92 unstimulated and 36.15 ± 2.88 stimulated: t(7) = 7.78, p < 0.001), as well as in the CA3 field (17.19 ± 1.12 unstimulated and 59.93 ± 6.22 stimulated: t(7) = 6.49, p < 0.001) (Fig. 4). These results indicate that long-term changes in synaptic efficiency (i.e., hippocampal long-lasting LTP) requires PARP-1 activity.
Figure 4.
The HFS protocol used for LTP induces an increase in PAR polymer in the hippocampus. The levels of PAR polymer were determined by immunohistochemistry in brains from mice killed 1 h after the application of six HFS stimulations to the entorhinal cortex (n = 3 mice per group). Bar chart illustrating the quantification of PAR polymer immunoreactivity in the CA1, CA3, and dentate gyrus (DG) of unstimulated (white bars) and stimulated (black bars) hippocampi. Significant differences of the data from unstimulated and stimulated slices for each hippocampal region. **p < 0.01.
Histone H1 is poly[ADP]-ribosylated and its expression decreases after OR training
The linker histone H1 is a bona fide _target of PARP-1 activation (Buki et al., 1995; Rouleau et al., 2004). To determine whether histone H1 is poly[ADP]-ribosylated during storage of the OR memory, hippocampal protein extracts from naive and trained mice obtained just after the training session were examined in bidimensional gels. The OR training session induced histone H1 poly[ADP]-ribosylation in the hippocampus, as shown by the acidic shift in the histone H1 pI (Fig. 5A). This effect was reverted when the training session was performed after Tiq-A administration. To quantify the change in histone H1 poly[ADP]-ribosylation during OR learning and memory, protein extracts, obtained 1 h after the training session from the hippocampus of naive and trained mice that were administered with Tiq-A or the vehicle alone, were analyzed in Western blots probed for PAR and histone H1 antibodies. OR training provoked a PARP-1-dependent increase in PAR-protein (two-way ANOVA, F(12,3) = 140.91, p < 0.001) and a decrease in histone H1 immunoreactivity (two-way ANOVA, F(12,3) = 86.03, p < 0.001) (Fig. 5B). Furthermore, when the same protein extracts were immunoprecipitated with the PAR polymer antibody and histone H1 was examined in Western blots, histone H1 was clearly poly[ADP]-ribosylated after OR training session in a PARP-1-dependent manner (two-way ANOVA, F(36,3) = 90.63, p < 0.001). Similar results were obtained when the extracts were immunoprecipitated with the anti-histone H1 antibody and the Western blots were probed for PAR polymer (two-way ANOVA, F(12,3) = 52.63, p < 0.001). We then evaluated the levels of histone H1 in the hippocampal CA1 field by immunohistochemistry 1 h after training (Fig. 5C). Untrained mice had more neurons with strong histone H1 immunoreactivity compared with trained mice (76.98 ± 3.51% for untrained mice and 30.57 ± 7.50% for trained mice; t(8) = 5.58, p < 0.001). This change in the pattern of histone H1 levels was completely abolished in the CA1 of mice trained in the presence of Tiq-A. Together, these results indicate that the exploration of new objects induces poly[ADP]-ribosylation of histone H1 and a decrease in histone H1 expression.
Figure 5.
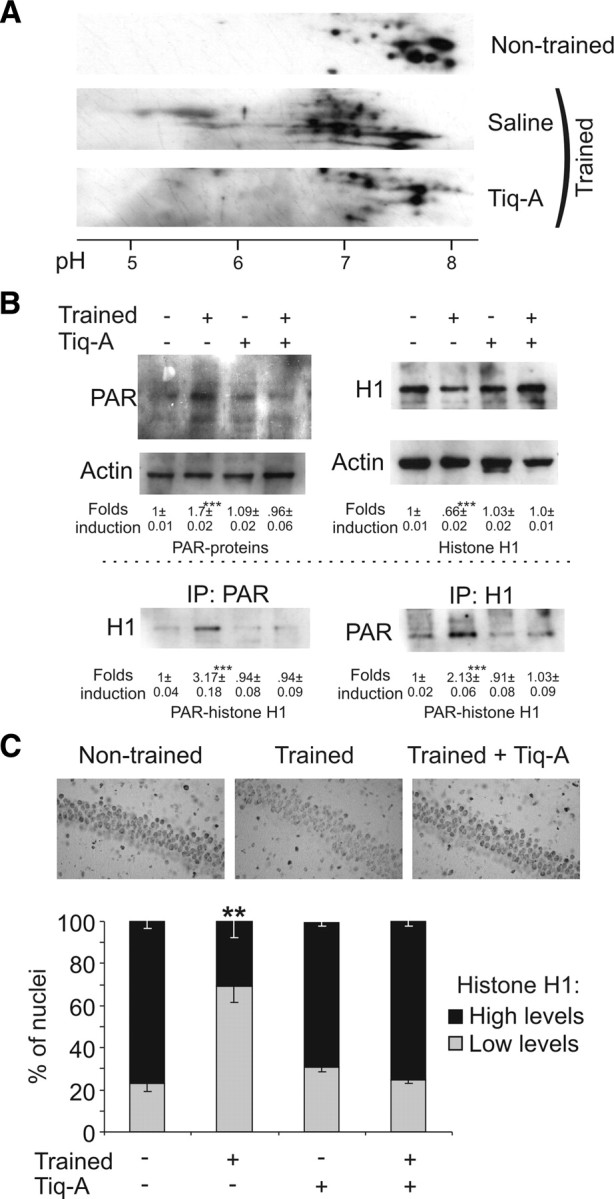
OR training sessions induced histone H1 poly[ADP]-ribosylation and protein decrease in the hippocampus. A, Protein extracts from the hippocampus obtained from untrained mice and those that had just finished their training, in the presence or absence of Tiq-A (0.5 mg/kg), were analyzed by two-dimensional electrophoresis, and histone H1 immunoreactivity was determined. Poly[ADP]-ribosylation of histone H1 is reflected by a shift in pI toward acidic pH values. B, Hippocampal protein extracts from untrained and trained mice that received Tiq-A (0.5 mg/kg) or the vehicle alone and were killed 15 min after training session were used to immunoprecipitate PAR polymer or histone H1. The immunoprecipitates were examined in Western blots probed for histone H1 and PAR polymer (bottom). As an internal control, 10% of the protein used for the immunoprecipitation assays was examined in Western blots probed for PAR polymer, histone H1, and actin (top). Values illustrate the densitometric quantification of PAR polymer and histone H1 immunoreactivity in each experimental group or the relative amount of histone H1 poly[ADP]-ribosylated (PAR-histone H1) in each experimental group with respect to the untrained animals. C, Immunohistochemistry of histone H1 expression in the hippocampal CA1 field of mice killed 1 h after training or manipulation in the presence or absence of Tiq-A. The top shows representative images of histone H1 in CA1 neurons after the manipulations. The bottom shows a bar graph of the percentage of neurons labeled with high or low amounts of histone H1 under the different conditions (n = 5 mice per groups). IP, Immunoprecipitation. **p < 0.01; ***p < 0.001.
PARP-1 regulates learning-induced gene expression by removing histone H1 from the promoters of cAMP response element-binding protein and nuclear factor-κB _target genes
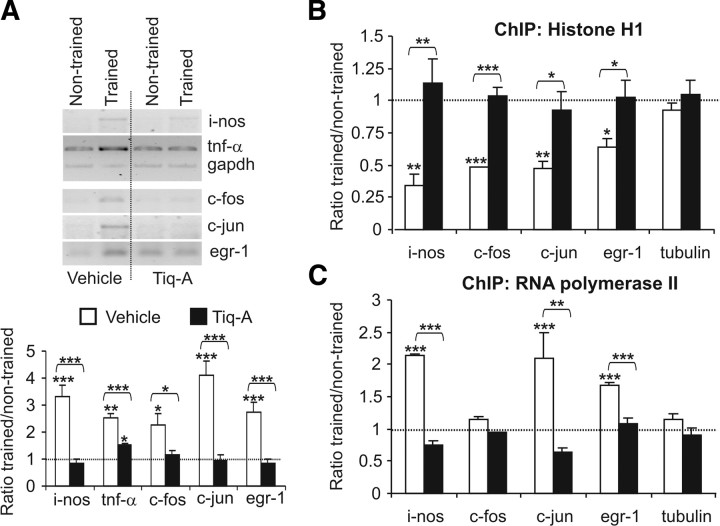
Histone H1 binds to nucleosome core particles near the entry/exit sites of linker DNA, influencing chromatin structure and gene regulation (Fan et al., 2005). It has been shown that learning consolidation requires the coordinated activation of the nuclear factor-κB (NF-κB) and cAMP response element-binding protein (CREB) transcription factors (Bozon et al., 2003; Ahn et al., 2008). The OR training session induced a twofold to fourfold increase in the expression of the NF-κB _target genes inducible nitric oxide synthase (i-nos) and tumor necrosis factor-α (tnf-α), and in the CREB _target genes c-fos, c-jun, and egr-1 in the hippocampus with respect to the untrained mice (for i-nos: 3.3 ± 0.42-fold induction, t(8) = 5.46, p < 0.001; for tnf-α: 2.51 ± 0.17-fold induction, t(8) = 5.37, p = 0.001; for c-fos: 2.41 ± 0.45-fold induction, t(16) = 2.6, p = 0.018; for c-jun: 4.09 ± 0.53-fold induction, t(16) = 3.68, p < 0.001; and for egr-1: 4.78 ± 0.38-fold induction, t(10) = 4.78, p < 0.001) (Fig. 6A). Interestingly, learning-induced NF-κB- and CREB-dependent transcription was blocked by the administration of Tiq-A before the training session.
Figure 6.
OR training promotes PARP-1-dependent histone H1 clearance and RNA pol II recruitment to NF-κB and CREB gene promoters. A, Semiquantitative reverse transcription-PCR analysis of i-nos, tnf-α, c-fos, c-jun, and egr-1 mRNA from the hippocampus of untrained mice and from that obtained 1 h after OR training in mice that received Tiq-A or the vehicle alone. GAPDH mRNA served as an internal control. The bar graph represents the trained/untrained ratio of the normalized values of gene expression in the presence or absence of Tiq-A (white and black bars, respectively; n = 5 mice per group). B, C, The levels of histone H1 (B) and RNA pol II (C) at the tnf-α, c-fos, c-jun, and egr-1 promoters were studied by chromatin immunoprecipitation in hippocampal chromatin complexes obtained from untrained and trained mice that received Tiq-A or the vehicle alone. Histograms represent the trained/untrained ratio of normalized values of gene promoter amplification in the presence or absence of Tiq-A (black and white bars, respectively; n = 3 mice per group). Asterisks above the line indicate a difference between the two treatments (vehicle and Tiq-A), whereas asterisks above a bar graph indicate a significant difference between trained and untrained mice. *p < 0.05; **p < 0.01; ***p < 0.001.
In many cases, transcriptional activation requiring histone H1 release may be mediated by poly[ADP]-ribosylation and the formation of the preinitiation complex, including RNA pol II (Kraus and Lis, 2003). To evaluate this hypothesis in learning and memory, we performed ChIP assays using histone H1 or RNA pol II antibodies on cross-linked chromatin extracts from the hippocampus of untrained or trained animals killed 1 h after training session having been exposed to Tiq-A or the vehicle alone (Fig. 6B,C). The OR exploration session induced a significant decrease of histone H1 associated with the i-nos, c-fos, c-jun, and egr-1 promoters in trained mice compared with untrained mice, as demonstrated by the trained/untrained promoter amplification ratio (ratio trained/untrained: for i-nos, 0.34 ± 0.09, t(6) = 4.66, p = 0.002; for c-fos, 0.48 ± 0.005, t(6) = 7.97, p < 0.001; for c-jun, 0.47 ± 0.06, t(6) = 3.85, p = 0.006; and for egr-1, 0.64 ± 0.066, t(6) = 2.95, p = 0.021) (Fig. 6B). In parallel, the OR training session induced a significant increase in the recruitment of RNA pol II to the NF-κB and CREB promoters studied (ratio trained/untrained: for i-nos, 2.15 ± 0.01, t(6) = 6.7, p < 0.001; for c-fos, 1.15 ± 0.03, t(6) = 1, p < 0.35; for c-jun, 2.1 ± 0.4, t(6) = 15.04, p < 0.001; and for egr-1, 1.68 ± 0.03, t(6) = 10.7, p < 0.001) (Fig. 6C). These changes were completely abolished when ChIP assays were performed with hippocampal chromatin extracts prepared from untrained and trained mice in the presence of Tiq-A. Interestingly, such effects were not found when similar ChIP experiments were performed on the tubulin promoter. Globally, these results indicate that PARP-1 activation induces local alterations in chromatin structure through histone H1 release and the recruitment of RNA pol II to gene promoters whose expression are required for memory consolidation.
Discussion
Epigenetics involves heritable and self-perpetuating modifications to DNA and chromatin (for review, see Surani et al., 2007), principally in the form of DNA methylation or histone modification. Epigenetic marking of the genome in metazoans is associated with the developmental processes of determination and differentiation, whereby totipotent stem cells are induced to terminally differentiate. Thus, epigenetic marking of the genome can be considered a persistent form of cellular memory by which terminally differentiated cells remember their phenotype. In the past 10 years, different laboratories have reported that histone modifications (acetylation and methylation) and DNA methylation are regulated in adult neurons in association with learning and memory processes (for review, see Roth and Sweatt, 2009; Gupta el al., 2010), as well as in response to seizure (Huang et al., 2002; Tsankova et al., 2004) and psychotropic drugs (Li et al., 2004; Kumar et al., 2005) and in mental disorders (Levenson and Sweatt, 2005). These data suggest that epigenetic regulation also have a critical role in the postmitotic cells function.
Cognitive processes require modifications in gene expression to consolidate information (Abel and Kandel, 1998; Milner et al., 1998; Abel and Lattal, 2001). Transcription regulation is a critical step in gene regulation, and many studies have focused on transcription factor activation (for review, see Alberini, 2009). However, some more recent studies have examined the role of changes in chromatin structure induced by the alterations to epigenetic factors during cognitive processes (Roth and Sweatt, 2009). Thus, histone acetylation and histone methylation have been demonstrated to be critical processes in regulating gene expression during learning and memory (Alarcón et al., 2004; Korzus et al., 2004; Levenson et al., 2004a; Wood et al., 2005; Vecsey et al., 2007; Fontán-Lozano et al., 2008; Gupta et al., 2010). Changes in DNA methylation during learning and memory processes have also been described as critical steps in gene expression (Miller et al., 2007; Lubin et al., 2008; Feng et al., 2010). Nevertheless, epigenetic factors such as poly[ADP]-ribosylation, sumoylation, and ubiquitination have been poorly studied in the cognitive context.
It was demonstrated recently that PARP-1 activity is induced under physiological conditions in the absence of DNA damage (Quénet et al., 2009). Moreover, PARP-1 activation has been shown to be required for long-term memory formation in Aplysia and mice (Cohen-Armon et al., 2004; Goldberg et al., 2009; Hernández et al., 2009), although the molecular mechanisms involved have not yet been described. Here we confirm that PARP-1 activity is needed in the mouse hippocampus to establish long-lasting synaptic plasticity events such as learning consolidation and long-lasting LTP. In memory consolidation, the temporal requirement of PARP-1 activity extended during the training session until 2 h after the end of the session. Similar results were obtained in Aplysia when PARP-1 inhibitor was administered before operant-conditioned training (Cohen-Armon et al., 2004). These data suggest that PARP-1 is involved in the changes of gene expression needed for long-lasting synaptic plasticity.
Like most forms of long-term synaptic plasticity (revised by Kandel, 2001), OR memory consolidation requires changes in gene expression (Rossato et al., 2007; Balderas et al., 2008; Romero-Granados et al., 2010). The impairment we detected in long-lasting synaptic and memory processes, together with an absence of any effect on such short-term events, and earlier data indicate that PARP-1 activity plays a critical role in regulating stimulus-induced changes in macromolecular synthesis required for long-term synaptic plasticity (Cohen-Armon et al., 2004; Goldberg et al., 2009). In Aplysia, PARP-1 activation is required for the transcription of ribosomal RNAs and ribosomal proteins (Hernández et al., 2009). Conversely, NF-κB- and CREB-dependent gene expression is required for learning and memory stabilization in mammals (Barco et al., 2002; Levenson et al., 2004b; Vecsey et al., 2007; Ahn et al., 2008; Fontán-Lozano et al., 2009) (for review, see Alberini, 2009). Our gene expression analysis in the hippocampus after OR training demonstrated the increase in NF-κB and CREB gene _targets and, in both cases, training-induced NF-κB- and CREB-dependent gene expression required of PARP-1 activation. It is well known that PARP-1 works as a coactivator of NF-κB in some cell types (Aguilar-Quesada et al., 2007), although to our knowledge this is the first description of PARP-1 and CREB transcriptional cooperation.
Gene expression during long-term synaptic plasticity requires the activation of transcription factors, as well as chromatin remodeling (Wolffe and Hayes, 1999; Rosenfeld and Glass, 2001). As a regulator of gene expression, PARP-1 seems to play a critical role in both these processes. Thus, PARP-1 contributes to NF-κB activation in the inflammatory response, provoking CREB binding protein recruitment independent of poly[ADP]-ribosylation (Hassa et al., 2005). In contrast, PARP-1 enzymatic activity is need for the recruitment of coactivators to Elk-1 and Hes-1 during neuronal activation and neurogenesis, respectively (Ju et al., 2004; Cohen-Armon et al., 2007). It has also been shown that PARP-1 contributes to chromatin remodeling and gene transcription in Drosophila under certain physiological conditions (Tulin and Spradling, 2003), probably by releasing histone H1 from specific promoters (Krishnakumar et al., 2008). Here we show that learning provokes an increase in the PAR polymer, and particularly in the poly[ADP]-ribosylation of histone H1, in brain regions relevant for learning and memory. This effect appears to promote local chromatin alterations at specific promoters attributable to histone H1 release, probably produced by degradation, and a decrease in its expression. Indeed, in mice lacking several of the genes that encode histone H1, there are alterations in chromatin structure and gene expression (Fan et al., 2005). Although the mechanism underlying the decrease in histone H1 expression remains unknown, we speculate that histone H1 may be a _target for caspases and/or the proteasome when activated after neuronal synaptic activity (Patrick, 2006; Kudryashova et al., 2009). Our results confirm that, under physiological conditions, memory formation induces chromatin alterations through histone H1 release from certain RNA polymerase II-specific, CREB- and NF-κB-dependent promoters. Interestingly, as indicated previously (Sandoval et al., 2004), we detected a decrease of histone H1 in the c-fos promoter but no change in RNA polymerase II recruitment. Hence, RNA polymerase II may be constitutively associated with the c-fos promoter, and for this reason c-fos can quickly increase its expression after stimulation. Thus, PARP-1 may be one of the many global chromatin-modifying activities that, like acetylases, deacetylases, and methylases, can also be specifically recruited to modify the structure of chromatin and its response to external signals.
Together, these data suggest a new mechanism that is active in memory consolidation, whereby activated PARP-1 molecules provoke poly[ADP]-ribosylation of histone H1 at specific DNA loci and probably that of other nuclear proteins, thereby affecting chromatin condensation. This expanded chromatin state may facilitate modifications in histone acetylation and/or histone and DNA methylation, to stabilize the information acquired in a genetic code.
Footnotes
This work was supported by the Agencia Espanola de Cooperacion Internacional, Junta de Andalucía Grant BIO-122, Direccion General de Investigacion Cientifica y Tecnica Grant BFU2008-01552, Instituto de Salud Carlos III (Red TerCel) Grant RD06/0010/0025, the Consejería de Salud (Fundación Progreso y Salud), and the Fundación Ramón Areces. We thank Dr. Sefton for critical reading and editorial assistance with the manuscript. We also thank Dr. Gómez-Skarmeta for critical reading of this manuscript and M. C. Sutil for technical assistance with animal handling.
References
- Abel T, Kandel E. Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res Rev. 1998;26:360–378. doi: 10.1016/s0165-0173(97)00050-7. [DOI] [PubMed] [Google Scholar]
- Abel T, Lattal KM. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr Opin Neurobiol. 2001;11:180–187. doi: 10.1016/s0959-4388(00)00194-x. [DOI] [PubMed] [Google Scholar]
- Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta-Leal A, Quiles-Pérez R, Rodríguez-Vargas JM, Ruiz de Almodóvar M, Conde C, Ruiz-Extremera A, Oliver FJ. Modulation of transcription by PARP-1: consequences in carcinogenesis and inflammation. Curr Med Chem. 2007;14:1179–1187. doi: 10.2174/092986707780597998. [DOI] [PubMed] [Google Scholar]
- Ahn HJ, Hernandez CM, Levenson JM, Lubin FD, Liou HC, Sweatt JD. c-Rel, an NF-kappaB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation. Learn Mem. 2008;15:539–549. doi: 10.1101/lm.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderas I, Rodriguez-Ortiz CJ, Salgado-Tonda P, Chavez-Hurtado J, McGaugh JL, Bermudez-Rattoni F. The consolidation of object and context recognition memory involve different regions of the temporal lobe. Learn Mem. 2008;15:618–624. doi: 10.1101/lm.1028008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, Kandel ER. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/s0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- Bozon B, Kelly A, Josselyn SA, Silva AJ, Davis S, Laroche S. MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Philos Trans R Soc Lond B Biol Sci. 2003;358:805–814. doi: 10.1098/rstb.2002.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MW, Aggleton JP. Recognition memory: What are the roles of the perirhinal cortex and hippocampus? Nat Rev Neurosci. 2001;2:51–61. doi: 10.1038/35049064. [DOI] [PubMed] [Google Scholar]
- Buki KG, Bauer PI, Hakam A, Kun E. Identification of domains of poly(ADP-ribose) polymerase for protein binding and self-association. J Biol Chem. 1995;270:3370–3377. doi: 10.1074/jbc.270.7.3370. [DOI] [PubMed] [Google Scholar]
- Clarke JR, Cammarota M, Gruart A, Izquierdo I, Delgado-García JM. Plastic modifications induced by object recognition memory processing. Proc Natl Acad Sci U S A. 2010;107:2652–2657. doi: 10.1073/pnas.0915059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Katzoff A, Levitan D, Susswein AJ, Klein R, Valbrun M, Schwartz JH. Long-term memory requires polyADP-ribosylation. Science. 2004;304:1820–1822. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Daoudal G, Debanne D. Long-term plasticity of intrinsic excitability: learning rules and mechanisms. Learn Mem. 2003;10:456–465. doi: 10.1101/lm.64103. [DOI] [PubMed] [Google Scholar]
- Dere E, Huston JP, De Souza Silva MA. The pharmacology, neuroanatomy and neurogenetics of the one-trial object recognition in rodent. Neurosci Biobehav Rev. 2007;31:673–704. doi: 10.1016/j.neubiorev.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontán-Lozano A, Sáez-Cassanelli JL, Inda MC, de los Santos-Arteaga M, Sierra-Domínguez SA, López-Lluch G, Delgado-García JM, Carrión AM. Caloric restriction increases learning consolidation and facilitates synaptic plasticity through mechanisms dependent on NR2B subunits of the NMDA receptor. J Neurosci. 2007;27:10185–10195. doi: 10.1523/JNEUROSCI.2757-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontán-Lozano A, Romero-Granados R, Troncoso J, Múnera A, Delgado-García JM, Carrión AM. Histone deacetylase inhibitors improve learning consolidation in young and in KA-induced-neurodegeneration and SAMP-8-mutant mice. Mol Cell Neurosci. 2008;39:193–201. doi: 10.1016/j.mcn.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Fontán-Lozano A, Romero-Granados R, del-Pozo-Martín Y, Suárez-Pereira I, Delgado-García JM, Penninger JM, Carrión AM. Lack of DREAM protein enhances learning and memory and slows brain aging. Curr Biol. 2009;19:54–60. doi: 10.1016/j.cub.2008.11.056. [DOI] [PubMed] [Google Scholar]
- Goldberg S, Visochek L, Giladi E, Gozes I, Cohen-Armon M. PolyADP-ribosylation is required for long-term memory formation in mammals. J Neurochem. 2009;111:72–79. doi: 10.1111/j.1471-4159.2009.06296.x. [DOI] [PubMed] [Google Scholar]
- Gruart A, Muñoz MD, Delgado-García JM. Involvement of the CA3–CA1 synapse in the acquisition of associative learning in behaving mice. J Neurosci. 2006;26:1077–1087. doi: 10.1523/JNEUROSCI.2834-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- Hernández AI, Wolk J, Hu JY, Liu J, Kurosu T, Schwartz JH, Schacher S. Poly-(ADP-ribose) polymerase-1 is necessary for long-term facilitation in Aplysia. J Neurosci. 2009;29:9553–9562. doi: 10.1523/JNEUROSCI.1512-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homburg S, Visochek L, Moran N, Dantzer F, Priel E, Asculai E, Schwartz D, Rotter V, Dekel N, Cohen-Armon M. A fast signal-induced activation of poly(ADP-ribose) polymerase: a novel downstream _target of phospholipase c. J Cell Biol. 2000;150:293–307. doi: 10.1083/jcb.150.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J Neurosci. 2002;22:8428–8522. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase is a critical component of memory formation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Kudryashova IV, Onufriev MV, Kudryashov IE, Gulyaeva NV. Caspase-3 activity in hippocampal slices reflects changes in synaptic plasticity. Neurosci Behav Physiol. 2009;39:13–20. doi: 10.1007/s11055-008-9089-z. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki T, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004a;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Choi S, Lee SY, Cao YA, Ahn HJ, Worley KC, Pizzi M, Liou HC, Sweatt JD. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. J Neurosci. 2004b;24:3933–3943. doi: 10.1523/JNEUROSCI.5646-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo Y, Schroeder FA, Youngs RM, Schmidt TW, Ferris C, Konradi C, Akbarian S. Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J Neurochem. 2004;90:1117–1131. doi: 10.1111/j.1471-4159.2004.02569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Milner B, Squire LR, Kandel ER. Cognitive neuroscience and the study of memory. Neuron. 1998;20:445–468. doi: 10.1016/s0896-6273(00)80987-3. [DOI] [PubMed] [Google Scholar]
- Patrick GN. Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Curr Opin Neurobiol. 2006;16:90–94. doi: 10.1016/j.conb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. London: Academic; 2001. [Google Scholar]
- Quénet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Romero-Granados R, Fontán-Lozano A, Delgado-García JM, Carrión AM. From learning to forgetting: behavioral, circuitry, and molecular properties define the different functional states of the recognition memory trace. Hippocampus. 2010;20:584–595. doi: 10.1002/hipo.20669. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- Rossato JI, Bevilaqua LR, Myskiw JC, Medina JH, Izquierdo I, Cammarota M. On the role of hippocampal protein synthesis in the consolidation and reconsolidation of object recognition memory. Learn Mem. 2007;14:36–46. doi: 10.1101/lm.422607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Aubin RA, Poirier GG. Poly(ADP-ribosyl)ated chromatin domains: access granted. J Cell Sci. 2004;117:815–825. doi: 10.1242/jcs.01080. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Rodríguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, López-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Squire LR, Zola-Morgan S. The medial temporal lobe memory system. Science. 1991;253:1380–1386. doi: 10.1126/science.1896849. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Kumar A, Nestler EJ. Histone modification at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 2004;24:5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly[ADP]-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virág L, Szabó C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- Visochek L, Steingart RA, Vulih-Shultzman I, Klein R, Priel E, Gozes I, Cohen-Armon M. PolyADP-ribosylation is involved in neurotrophic activity. J Neurosci. 2005;25:7420–7428. doi: 10.1523/JNEUROSCI.0333-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modifications. Nucleic Acid Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficit in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]