Abstract
Hepatitis B virus (HBV) infections rely on the proper functioning of the viral polymerase enzyme, a specialized reverse transcriptase (RT) with multiple activities. All currently approved antiviral drugs for the treatment of chronic hepatitis B virus, except for interferon, _target the RT and belong to the same chemical class - they are all nucleoside analogs. Viral DNA synthesis is carried out by the RT enzyme in several different steps, each with distinct RT conformational requirements. In principle, each stage may be _targeted by distinct antiviral drugs. In particular, the HBV RT has the unique ability to initiate viral DNA synthesis using itself as a protein primer in a novel protein priming reaction. In order to help identify RT inhibitors and study their mechanisms of action, a number of experimental systems have been developed, each varying in its ability to dissect the protein priming and the subsequent stages of viral DNA synthesis reaction at the molecular level. Two of the most effective drugs to date, entecavir and tenofovir, can inhibit both the protein priming and the subsequent DNA elongation stages of HBV DNA synthesis. Interestingly, clevudine, a thymidine analog, can inhibit both protein priming and DNA elongation in a non-competitive manner and without being incorporated into the viral DNA. Thus, a nucleoside RT inhibitor (NRTI) can functionally mimic a non-NRTI (NNRTI) in its inhibition of the HBV RT. Therefore, novel NRTIs as well as NNRTIs may be developed to inhibit the DNA synthesis activity of the HBV RT. Furthermore, additional activities of the RT that are also essential to HBV replication, including specific recognition of the viral RNA and its packaging into viral nucleocapsids, may be exploited for antiviral development. To achieve a more potent inhibition of viral replication and ultimately cure chronic HBV infection, the next generation of anti-HBV therapies will likely need to include NRTIs, NNRTIs, and other agents that _target the viral RT as well as other viral and host factors in various combinations. This article forms part of a symposium in Antiviral Research on "An unfinished story: from the discovery of the Australia antigen to the development of new curative therapies for hepatitis B."
Reverse transcriptase structure and functions
Chronic hepatitis B virus (HBV) infection is a worldwide health problem, affecting over 350 million people and resulting in about a million deaths per year (El-Serag, 2012). The infectious agent is a viral particle, consisting of a nucleocapsid surrounded by a lipid envelope studded with viral envelope proteins. Inside the nucleocapsid is a relaxed circular DNA (rcDNA) genome. The viral polymerase, a reverse transcriptase (RT), is encoded in the largest open reading frame on the viral genome. The coding sequence of the RT gene alone is 2.5kb, though the entire viral genome is a mere 3.2 kb. Every other HBV gene overlaps with the polymerase (Hu and Seeger, 2015).
During an infection, the virus enters a cell when surface proteins bind to a recently identified liver-specific receptor (Yan et al., 2012). Inside the cell, the rcDNA genome must be delivered to the host cell nucleus. The rcDNA is then repaired by removal of the polymerase protein, which is covalently attached to the rcDNA, and by closure of the two strands. This forms the covalently closed circular DNA (cccDNA) which is the source of viral mRNA transcripts (Hu and Seeger, 2015). The largest viral mRNA transcript, the pre-genomic RNA (pgRNA), encodes viral polymerase and capsid proteins and additionally serves as the template for viral DNA synthesis (Hu and Seeger, 2015; Seeger et al., 2007; Summers and Mason, 1982). The pgRNA is degraded during synthesis of rcDNA due to the polymerase’s RNase H activity, which is discussed in a companion article in this series (Tavis and Lomonosova, 2015).
Synthesis of the viral genome is accomplished in several steps by the HBV polymerase, and importantly, the DNA is synthesized inside a newly formed nucleocapsid (Hu and Seeger, 2015). The first step occurs when the RT associates with the pgRNA and together they are packaged (or encapsidated) into an assembling capsid. This RNA binding occurs at the RNA packaging signal, called epsilon (ε RNA), which is a stem-loop structure on the 5’ end of the pgRNA (Figure 1A). This ε RNA binding by the RT is dependent on the presence of host cell chaperones which form a complex with the RT (Hu and Anselmo, 2000; Hu et al., 2004; Hu and Seeger, 1996b; Hu et al., 1997; Stahl et al., 2007b).
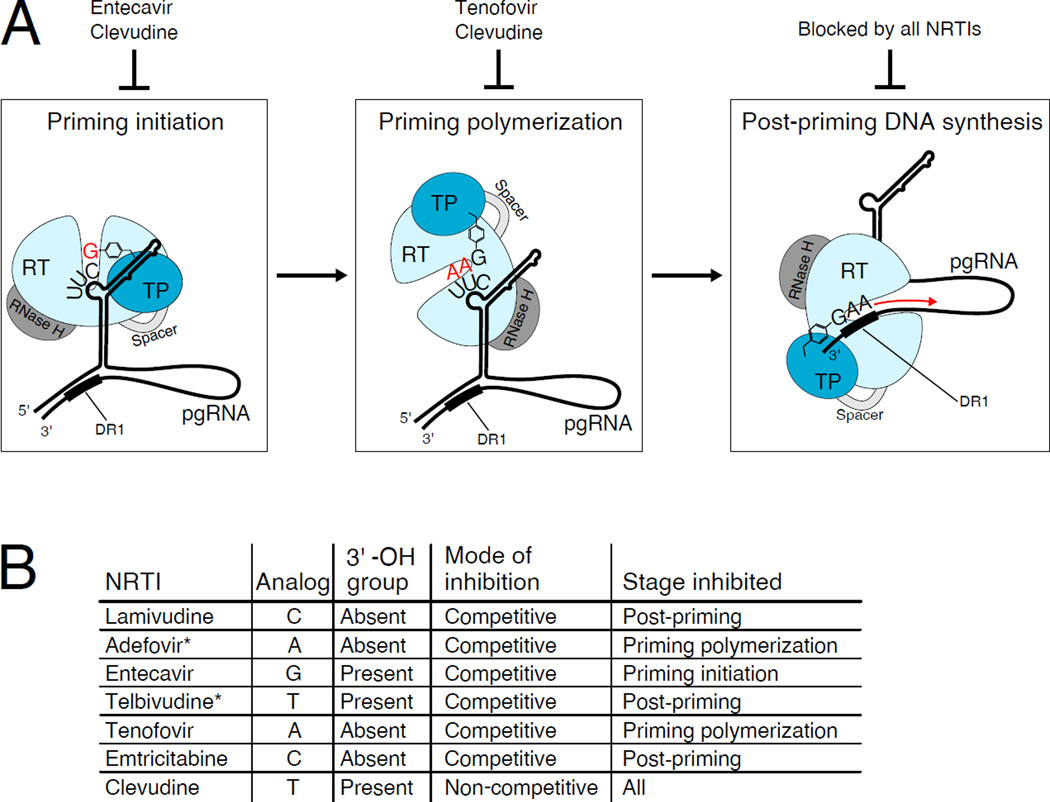
Figure 1. Inhibitors block the early stages of DNA synthesis by hepatitis B virus.
A. The hepatitis B virus (HBV) polymerase consists of a terminal protein domain (TP), a spacer domain, the reverse transcriptase domain (RT), and an RNase H domain. DNA synthesis begins after the polymerase binds to the viral pre-genomic RNA (pgRNA), and the subsequent steps of DNA synthesis are performed inside the nucleocapsid (not shown). Also, the polymerase is also associated with host chaperone proteins (not shown). Early DNA synthesis is performed in three stages, each of which has distinct requirements for polymerase conformation and the nucleotide used. First, initiation of protein priming occurs, which attaches a deoxyguanosine nucleotide (red “G”) to a tyrosine residue (shown as an aromatic ring) on the TP domain. Next, priming continues to polymerize two deoxyadenosine nucleotides (red “AA”). Once this short oligonucleotide is synthesized, it is translocated and anneals to the 3’ end of the pgRNA at a direct repeat 1 (DR1) sequence, and the minus strand of DNA continues to elongate (red arrow). Each of these stages can be blocked by nucleoside RT inhibitors (NRTI). B. NRTIs which _target HBV polymerase are listed, along with information about their chemical structure and mode of action (De Clercq et al., 2010). * It is assumed, but has not been determined if these NRTIs act competitively and only affect the indicated stage of DNA synthesis.
On the pgRNA, the ε RNA hairpin contains an internal bulge which is necessary for the first step in DNA synthesis, called protein priming (see Figure 1A). Located on the internal bulge is a 5’-UUC-3’ sequence, which is the template for initial synthesis of an oligonucleotide with the sequence 5’-dGAA-3’ (Jones et al., 2012; Nassal and Rieger, 1996; Wang and Seeger, 1993). The requirement for a primer is met by the HBV RT protein itself. In a biologically rare occurrence, a tyrosine residue on the RT acts as a primer, where its free -OH group covalently links to the initiating nucleotide, dGMP. This first step in protein priming is referred to as priming initiation, and the addition of the subsequent adenine nucleotides is called priming polymerization (Jones et al., 2012; Wang and Hu, 2002). The product of protein priming is therefore a short DNA oligonucleotide, 5’-dGAA-3’, which is covalently attached to the RT protein through a phosphotyrosyl bond. Subsequently, this polymerase-nascent DNA complex is translocated to a direct repeat 1 (DR1) sequence near the 3’ end of the pgRNA to continue minus strand synthesis (Figure 1A). Each of these steps is thought to require a distinct conformational state of the RT (Jones et al., 2014; Lin et al., 2008; Stahl et al., 2007a; Wang and Hu, 2002).
Clues into the phenomenon of protein priming came early on when a large protein was found to be covalently attached to the viral rcDNA genome, and was even found attached to the smallest detectable growing strands of DNA when using duck HBV (DHBV) (Gerlich and Robinson, 1980; Molnar-Kimber et al., 1983). Several years later, the identification of the attached protein as the polymerase led to the implication that all necessary activities for rcDNA synthesis—primer, polymerase, and RNase H—were contained in a single protein (Bartenschlager and Schaller, 1988; Wang and Seeger, 1992). The polymerase has four domains which facilitate these principal roles. Beginning at the N-terminus, these domains are the terminal protein domain (TP) which contains the tyrosine primer (Zoulim and Seeger, 1994), followed by a spacer domain, the RT domain which is responsible for catalyzing DNA synthesis, and the RNase H domain which is critical for degrading the pgRNA (Figure 1A).
The protein priming step is followed by the synthesis of the complete minus strand DNA, during which the pgRNA is degraded by the RNase H activity of the RT. Then the plus strand of rcDNA is generated using the minus strand as the template (Hu and Seeger, 1996a; Summers and Mason, 1982). Thus, like other RTs, the HBV polymerase uses both RNA and DNA as templates to synthesize its dsDNA genome.
It should be no surprise that the polymerase enzyme is the main _target of antiviral drugs (Clark and Hu, 2015; Jones and Hu, 2013a). It is the only viral protein with enzymatic activity, and the priming, DNA synthesis, and RNase H functions are all critical to the viral replication cycle. Although much is inferred about the structure of the HBV RT, there is yet no high-resolution structure of this important enzyme. Therefore, structural predictions, including drug interactions, are based on the well-studied RT of the human immunodeficiency virus (HIV), for which several crystal structures have been published. The HIV and HBV RT have approximately 20% similarity in the RT domain (Das et al., 2001), and 33% similarity in the RNase H domain (Tavis et al., 2013). Owing to the similarity of the HBV RT to that of HIV, several NRTIs are active against both RTs. However, the TP and spacer domains of the HBV polymerase are unique and have no structural homologs.
Current antivirals _target HBV DNA synthesis
Current treatments for chronic HBV infection have been discussed in other issues of this compendium (Block et al., 2015; Chang and Guo, 2015; Cheng et al., 2015; Gish et al., 2015a; Gish et al., 2015b; Tavis and Lomonosova, 2015; Yan et al., 2015; Zlotnick et al., 2015). They include interferon and nucleoside RT inhibitors (NRTIs, also called nucleotide RT inhibitors if they are the phosphorylated form, such as tenofovir and adefovir) (De Clercq et al., 2010; Scaglione and Lok, 2012). Interferon therapy is effective only in a minority of patients and is associated with side effects and toxicities. Five NRTIs have been approved by the United States Food and Drug Administration (FDA) for HBV treatment, namely lamivudine, adefovir, entecavir, telbivudine, and tenofovir (which are respectively analogs of C, A, G, T, and A) (Figure 1B). In addition, the cytidine analog emtricitabine is FDA approved for use against HIV, and known to also suppress HBV during treatment of HIV/HBV co-infected patients (Cui et al., 2015).
Clevudine is a thymidine analog approved for HBV treatment in the Philippines and South Korea (Balakrishna Pai et al., 1996; Chu et al., 1995; Jang et al., 2011; Korba et al., 2006). It was shown, after cessation of treatment, to maintain suppression of HBV replication over 6 months (Lee et al., 2006; Peek et al., 2001), very unusual for a polymerase inhibitor. Also different from many other NRTIs, clevudine is not considered an obligate chain terminator because of the presence of a 3’-OH group (Korba et al., 2006). Interestingly, in studies with Epstein-Barr virus, clevudine inhibited the DNA polymerase without being incorporated into the DNA (Yao et al., 1996), a finding which was later reproduced in tests with HBV (see below). Due to drug resistance and myopathy in a small proportion of patients (Kim et al., 2012; Seok et al., 2009), clevudine is being tested at lower doses and in combination with adefovir, which did not exhibit resistance after 96 weeks (Tak et al., 2014).
NRTIs suppress HBV replication but rarely provide a cure. The potentially life-long treatment regimen can thus be associated with drug resistance or toxicity (Scaglione and Lok, 2012; Zoulim and Locarnini, 2009). Therefore, there remains a need to identify novel compounds that can inhibit the RT or other viral _targets and for convenient screening systems to facilitate their identification.
Insights into polymerase function through the development of cell-free assays
Over the years, several experimental systems have emerged for studying the molecular biology of the HBV polymerase using different strategies. Early protein priming experiments were first developed using DHBV. With this system, it was found that priming requires the polymerase, ε RNA, and host cell chaperone proteins like the Hsp90 complex (Hu and Anselmo, 2000; Hu and Seeger, 1996b; Hu et al., 2002; Hu et al., 1997; Wang and Seeger, 1992). Similar to what was later found in HBV, the DHBV polymerase uses a conserved tyrosine residue to act as a primer (Y63 for HBV and Y96 for DHBV) (Lanford et al., 1997; Weber et al., 1994; Zoulim and Seeger, 1994). This system also gave insights into alternate sites used in protein priming in vitro. Priming at other residues can occur in vitro including other tyrosine and serine residues on both the TP and RT domains (Beck and Nassal, 2011; Boregowda et al., 2011). Also, the TP domain, normally attached to the RT domain through the spacer, can be expressed as a two-component system with the TP and RT domains as separate fragments, which are still priming active when combined (i.e. trans complementation). This facilitated truncation of the fragments to define minimally important sequences for priming and ε RNA binding (Boregowda et al., 2012). Cell-free systems using DHBV could be useful for drug screening, however, there is the obvious caveat of incongruent effects between the duck and human viruses.
Efforts to develop similar priming assays in vitro for the human HBV polymerase were unsuccessful for many years due to difficulties in purifying a functionally active polymerase in large enough quantities for testing. Also several systems were developed but were unable to reproduce the in vivo requirements, such as the ε RNA dependence. For example, expression of human HBV RT in insect cells using a recombinant baculovirus system shows in vitro priming at low levels and uses the authentic Y63 priming residue, but protein priming is ε RNA-independent (Lanford et al., 1995; Lanford et al., 1997). Another in vitro system is HBV polymerase expressed in E. coli and reconstituted with chaperone proteins. This system exhibits ε RNA binding activity, but is not competent in protein priming. RNA binding requirements of both the ε RNA and the RT were mapped using this system (Hu and Anselmo, 2000; Hu and Boyer, 2006; Hu et al., 2004). These systems have their utility, but cannot recapitulate each step performed by the RT.
Addressing most of these shortcomings, a full-length human HBV polymerase was expressed and purified using a mammalian cell culture system (Jones et al., 2012). Because the polymerase and ε RNA are co-expressed and then purified together from human cells, the natural host chaperones are attached and need not be added exogenously. This system allows in vitro testing for ε RNA binding and the ε-dependent protein priming activity because the reaction is ε RNA dependent (Jones et al., 2012). Protein priming in vitro can be studied in detail with this system, including the ability to separately analyze both priming initiation (attaching dGMP to Y63 in the TP domain) and priming polymerization (addition of two dAMP nucleotides to the initiating nascent DNA). Furthermore, this system is capable of both drug screening and the evaluation of currently used compounds to elucidate their exact molecular mechanisms.
Interactions between RT and NRTI by competitive and non-competitive means
Competitive inhibition has been the assumed mode of action for all NRTIs. The polymerase that normally recognizes nucleotide triphosphates will instead recognize the triphosphate form of the inhibitor in a competitive manner. The nucleotide analog is thus incorporated into the DNA strand. Several NRTIs lack a 3’ -OH group, which prevents bridging to the next incoming nucleotide, and thus DNA synthesis is terminated. If an inhibitor does have a 3’ -OH, it usually prevents elongation only after several more bases are added due to a delayed chain termination effect (De Clercq, 2010; De Clercq et al., 2010; Sarafianos et al., 2009). The compound ribavirin is an exception, acting as a mimic of both adenine and guanosine on RNA, but not terminating synthesis—instead it induces mutations when a ribavirin-containing strand is used as template for a new strand of RNA (Crotty et al., 2002).
One consideration in regards to polymerase inhibition is the multiple conformational states adopted by the HBV polymerase in order to carry out its multiple functions (see Figure 1). Each stage—priming initiation (requiring dGTP), priming polymerization (requiring dATP), and subsequent DNA synthesis (requiring all nucleotides)—has a distinct conformational requirement from the RT, and thus may be _targeted individually (Jones et al., 2012; Jones and Hu, 2013b; Lin et al., 2008; Staschke and Colacino, 1994; Wang and Seeger, 1992; Wang and Hu, 2002). This conformation-specific inhibition can be through the NRTI which mimics that stage’s nucleotide requirement, however non-NRTIs may be found which are stage-specific as well. For example, a non-NRTI compound may be found which inhibits the polymerase while in a conformation specific to the priming initiation step, but may not inhibit during the priming polymerization step due to differences in polymerase conformation.
Antiviral effects would presumably be the strongest when directed against early events in viral replication, such as priming. The sole approved drug which _targets priming initiation, entecavir, is indeed one of the most effective. It is a guanosine analog and acts competitively to inhibit DNA synthesis. Entecavir was shown to be incorporated into the DNA strand. It has a 3’ -OH, and thus does not immediately terminate synthesis, but instead ceases elongation in a delayed fashion. Specifically, the RT pauses at the third nucleotide after entecavir is incorporated. This occurs because the 3’ end, where a new nucleotide would be attached, is repelled from the active site when the growing strand includes entecavir (Jones et al., 2013; Seifer et al., 1998; Tchesnokov et al., 2008). Emtricitabine, lamivudine, and tenofovir also act competitively. They are incorporated into the DNA strand and, lacking a 3’ -OH, directly prevent chain elongation (De Clercq et al., 2010; Scaglione and Lok, 2012). Tenofovir, an adenosine analog, was shown to specifically inhibit priming polymerization, when two dAMPs are normally added, also in a competitive manner (Jones et al., 2013).
Surprisingly, a novel mechanism for polymerase inhibition emerged recently—an NRTI that acts non-competitively. Clevudine is a thymidine analog and thus should not inhibit priming initiation (which requires dGTP) or priming polymerization (which requires dATP), but instead should only block downstream DNA synthesis after priming. However, it inhibits all stages of DNA synthesis, and it does so non-competitively and without being incorporated into the DNA strand. Thus, although a nucleoside analog, the same chemical class as all other inhibitors of HBV RT, the mode of action of clevudine is distinct (see Figure 1) (Jones et al., 2013).
Although its exact polymerase binding site is not known, clevudine is thought to inhibit the HBV polymerase by binding to the nucleotide triphosphate binding pocket within the RT domain, just like other NRTIs. This was evidenced by the finding that only clevudine triphosphate, and not the mono- or diphosphate forms could inhibit the RT (Jones et al., 2013). Also clevudine-resistant mutations have been found in the dNTP binding site (Kwon et al., 2010). Interestingly, these clevudine-resistant polymerase mutants were also resistant to lamivudine and emtricitabine (both cytidine analogs), and to telbivudine (a thymidine analog) demonstrating shared constraints (De Clercq et al., 2010; Kwon et al., 2010).
Despite this evidence that clevudine may bind at the dNTP binding pocket of the HBV RT, clevudine is somehow not incorporated into the DNA strand. This may be due to unfavorable conformational constraints, according to one study which modeled clevudine binding to the active site (Chong and Chu, 2002). Since clevudine is not incorporated into the DNA chain, it could potentially bind an allosteric site close to the active site, altering the conformation of the active site to prevent DNA synthesis, i.e., an NRTI actually functions as a non-NRTI (NNRTI). Supporting this hypothesis, clevudine was found to act synergistically to inhibit HBV replication when combined with entecavir, lamivudine, adefovir or tenofovir (Niu et al., 2010), suggesting its ability to act cooperatively with other NRTIs. Interestingly, when combining clevudine and telbivudine (both thymidine analogs) an antagonistic effect was observed. It was proposed that this did not relate to any interactions with the polymerase itself, but instead that due to their chemical similarity they compete for other enzymes which are responsible for their uptake and phosphorylation (Niu et al., 2010).
Clevudine was shown to inhibit all stages of DNA synthesis when tested with the human-cell purified HBV polymerase described above. It blocks the initial priming reaction which requires dGTP, despite the fact that clevudine is not a guanosine analog. This finding was reinforced when the native 5’-UUC-3’ template sequence found in the internal bulge on the ε RNA was mutated to 5’-UUG-3’ or 5’-UUA-3’. These mutated templates thus require dCTP or TTP, however clevudine inhibited priming initiation in all cases (Jones et al., 2013). The next step, priming polymerization, was also inhibited by clevudine. Similar to tenofovir, an adenosine analog, clevudine blocked priming polymerization (which requires dATP) despite clevudine being a thymidine analog (Jones et al., 2013). Finally, clevudine can block, in a non-competitive manner, the post-priming DNA synthesis stage which forms the completed genome. During none of its inhibitory activity is clevudine incorporated into the DNA (Jones et al., 2013).
Potential for developing novel RT inhibitors, in combination with current NRTIs, for more complete inhibition of HBV replication and to cure chronic infection
Clevudine’s novel non-competitive inhibition provides an exciting proof of principle—NRTIs can act as non-NRTIs. This would be a welcome addition to the arsenal of compounds currently employed to treat HBV, which all use the same mechanism of action. When NRTIs and non-NRTIs are used together, their combinatorial effect can be synergistic in reducing viral load and provide a higher threshold to viral resistance. NNRTIs have been used against HIV for several years and bind to a pocket near the active site (Das et al., 2012; Sarafianos et al., 2009). This allosteric site, when bound to non-NRTIs, prevents the conformational changes required for the HIV RT to affix a new nucleotide. Unfortunately, anti-HIV non-NRTIs display no cross-reactivity to HBV.
Despite several attempts to devise a combination therapy that is more effective than single-compound, all current indications for HBV treatment are for monotherapy (Hadziyannis et al., 2013; Zhu et al., 2009). This is likely due to the single mode of inhibition for the components in the combined therapy trials. However, one study showed a synergistic effect when clevudine was combined with other NRTIs, as measured by viral DNA reduction (Niu et al., 2010). A recent clinical trial tested clevudine alone versus clevudine combined with adefovir. Only the combination therapy showed no resistance, and the decrease in viral load was stronger in the combined therapy (Tak et al., 2014). The most promising developments may come when derivatives of clevudine without its toxic effects or similarly acting non-NRTI compounds are used in combination with approved NRTIs, as is the case with HIV. In the meantime, NRTIs will continue to be a mainstay of anti-HBV therapy. Several new NRTIs are in development including variations of tenofovir and adefovir, and the novel compounds besifovir, elvucitabine, lagociclovir valactate, and valtorcitabine (Menendez-Arias et al., 2014; Yuen et al., 2015).
Other compounds which inhibit the HBV polymerase have been found, but none are in clinical trials. Hemin and other porphyrin compounds strongly inhibited the binding of HBV polymerase to ε RNA (Lin and Hu, 2008). A carbonyl J acid derivative, KM-1, _targets the binding of template and primer to the HIV polymerase (Skillman et al., 2002; Wang et al., 2004), and was found to block the binding of ε RNA to HBV polymerase as well (Wang et al., 2012). As polymerase-ε RNA interaction is a prerequisite for not only protein priming, but also pgRNA packaging, drugs _targeting this early interaction can be expected to be highly potent. Another critical feature of the HBV polymerase enzyme is its RNase H activity. Not only essential for degrading the pgRNA, this domain also contains several residues essential for DNA synthesis (Tavis and Lomonosova, 2015). The ability to screen compounds which are inhibitory to this domain has advanced recently, and β-thujaplicinol and HID compounds were shown to have anti-RNase H activity (Cai et al., 2014; Hu et al., 2013; Tavis et al., 2013). Additionally, a method to inhibit the HBV replication cycle would be to _target host chaperone proteins that facilitate HBV RT function (Hu et al., 2004; Hu and Seeger, 1996b).
The burden of morbidity and mortality due to chronic HBV infection has decreased since the introduction of polymerase-directed antiviral drugs, and the potential for finding new HBV RT inhibitors is high. While the polymerase, with its many functions, has been successfully _targeted by NRTIs, the advent of combination therapies which include non-NRTIs as well as inhibitors of other viral components should further increase the effectiveness of HBV drug therapy. There are many strategies and compounds in development which _target HBV or its life cycle beyond the RT and are further discussed elsewhere in this symposium (Block et al., 2015; Chang and Guo, 2015; Cheng et al., 2015; Gish et al., 2015a; Gish et al., 2015b; Tavis and Lomonosova, 2015; Yan et al., 2015; Zlotnick et al., 2015). Assuming that new cccDNA formation is needed to sustain HBV persistence, combination therapies sufficiently potent to completely block rcDNA synthesis should be able to clear cccDNA and thus cure chronic HBV infection, once the preexisting cccDNA is removed by the host (Hu and Seeger, 2015).
Highlights.
The HBV RT, unique in its function as a protein primer for DNA synthesis, is the main _target of current antiviral therapy.
Recently developed experimental systems have helped to reveal the molecular mechanism of RT inhibitors.
Clevudine is a novel NRTI against HBV which acts non-competitively and is not incorporated into viral DNA.
NRTIs and NNRTIs, and agents which _target other viral or host factors may be combined in future therapies.
Acknowledgements
This work was supported by a Public Health Service grant (R01 AI043453to J.H.) and a National Research Service Award grant (T32 CA 60395 to D.C.) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balakrishna Pai S, Liu SH, Zhu YL, Chu CK, Cheng YC. Inhibition of hepatitis B virus by a novel L-nucleoside, 2'-fluoro-5-methyl-beta-L-arabinofuranosyl uracil. Antimicrob Agents Chemother. 1996;40:380–386. doi: 10.1128/aac.40.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. Embo J. 1988;7:4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Nassal M. A Tyr residue in the reverse transcriptase domain can mimic the protein-priming Tyr residue in the terminal protein domain of a hepadnavirus P protein. J Virol. 2011;85:7742–7753. doi: 10.1128/JVI.00482-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Rawat S, Brosgart CL. Chronic hepatitis B: A wave of new therapies on the horizon. Antiviral Res. 2015;121:69–81. doi: 10.1016/j.antiviral.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boregowda R, Adams C, Hu J. TP-RT Domain Interactions of Duck Hepatitis B Virus Reverse Transcriptase In Cis and In Trans during Protein-Primed Initiation of DNA Synthesis In Vitro. J Virol. 2012;86:6522–6536. doi: 10.1128/JVI.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boregowda RK, Lin L, Zhu Q, Tian F, Hu J. Cryptic protein priming sites in two different domains of duck hepatitis B virus reverse transcriptase for initiating DNA synthesis in vitro. J Virol. 2011;85:7754–7765. doi: 10.1128/JVI.00483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CW, Lomonosova E, Moran EA, Cheng X, Patel KB, Bailly F, Cotelle P, Meyers MJ, Tavis JE. Hepatitis B virus replication is blocked by a 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) inhibitor of the viral ribonuclease H activity. Antiviral Res. 2014;108:48–55. doi: 10.1016/j.antiviral.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Guo JT. Treatment of chronic hepatitis B with pattern recognition receptor agonists: Current status and potential for a cure. Antiviral Res. 2015 doi: 10.1016/j.antiviral.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Li F, Bility MT, Murphy CM, Su L. Modeling hepatitis B virus infection, immunopathology and therapy in mice. Antiviral Res. 2015;121:1–8. doi: 10.1016/j.antiviral.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y, Chu CK. Understanding the unique mechanism of L-FMAU (clevudine) against hepatitis B virus: molecular dynamics studies. Bioorg Med Chem Lett. 2002;12:3459–3462. doi: 10.1016/s0960-894x(02)00747-3. [DOI] [PubMed] [Google Scholar]
- Chu CK, Ma T, Shanmuganathan K, Wang C, Xiang Y, Pai SB, Yao GQ, Sommadossi JP, Cheng YC. Use of 2'-fluoro-5-methyl-beta-L-arabinofuranosyluracil as a novel antiviral agent for hepatitis B virus and Epstein-Barr virus. Antimicrob Agents Chemother. 1995;39:979–981. doi: 10.1128/aac.39.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DN, Hu J. Unveiling the roles of HBV polymerase for new antiviral strategies. Future virology. 2015;10:283–295. doi: 10.2217/fvl.14.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Cameron C, Andino R. Ribavirin's antiviral mechanism of action: lethal mutagenesis? J Mol Med (Berl) 2002;80:86–95. doi: 10.1007/s00109-001-0308-0. [DOI] [PubMed] [Google Scholar]
- Cui G, Xu X, Diao H. Comparative Meta-Analysis of Tenofovir Disoproxil Fumarate versus Emtricitabine and Tenofovir Disoproxil Fumarate as Treatments for Patients with Chronic Hepatitis B. Scientific reports. 2015;5:11854. doi: 10.1038/srep11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Martinez SE, Bauman JD, Arnold E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol. 2012;19:253–259. doi: 10.1038/nsmb.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, Arnold E. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC) J Virol. 2001;75:4771–4779. doi: 10.1128/JVI.75.10.4771-4779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. In search of a selective therapy of viral infections. Antiviral Res. 2010;85:19–24. doi: 10.1016/j.antiviral.2009.10.005. [DOI] [PubMed] [Google Scholar]
- De Clercq E, Ferir G, Kaptein S, Neyts J. Antiviral treatment of chronic hepatitis B virus (HBV) infections. Viruses. 2010;2:1279–1305. doi: 10.3390/v2061279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich WH, Robinson WS. Hepatitis B virus contains protein attached to the 5' terminus of its complete DNA strand. Cell. 1980;21:801–809. doi: 10.1016/0092-8674(80)90443-2. [DOI] [PubMed] [Google Scholar]
- Gish RG, Given BD, Lai CL, Locarnini SA, Lau JY, Lewis DL, Schluep T. Chronic hepatitis B: Virology, natural history, current management and a glimpse at future opportunities. Antiviral Res. 2015a;121:47–58. doi: 10.1016/j.antiviral.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Gish RG, Yuen MF, Chan HL, Given BD, Lai CL, Locarnini SA, Lau JY, Wooddell CI, Schluep T, Lewis DL. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015b;121:97–108. doi: 10.1016/j.antiviral.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Hadziyannis SJ, Vassilopoulos D, Hadziyannis E. Chapter Seven - The Natural Course of Chronic Hepatitis B Virus Infection and Its Management. In: Erik De C, editor. Advances in Pharmacology. Academic Press; 2013. pp. 247–291. [DOI] [PubMed] [Google Scholar]
- Hu J, Anselmo D. In Vitro Reconstitution of a Functional Duck Hepatitis B Virus Reverse Transcriptase: Posttranslational Activation by Hsp90. J Virol. 2000;74:11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol. 2004;78:13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Seeger C. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 1996a;275:195–208. doi: 10.1016/s0076-6879(96)75013-9. [DOI] [PubMed] [Google Scholar]
- Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proceedings of the National Academy of Sciences of the United States of America. 1996b;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Seeger C. Hepadnavirus Genome Replication and Persistence. Cold Spring Harbor Perspectives in Medicine. 2015;5 doi: 10.1101/cshperspect.a021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Toft D, Anselmo D, Wang X. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J Virol. 2002;76:269–279. doi: 10.1128/JVI.76.1.269-279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. Embo J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Cheng X, Cao F, Huang A, Tavis JE. beta-Thujaplicinol inhibits hepatitis B virus replication by blocking the viral ribonuclease H activity. Antiviral Res. 2013;99:221–229. doi: 10.1016/j.antiviral.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Jang JH, Kim JW, Jeong SH, Myung HJ, Kim HS, Park YS, Lee SH, Hwang JH, Kim N, Lee DH. Clevudine for chronic hepatitis B: antiviral response, predictors of response, and development of myopathy. Journal of viral hepatitis. 2011;18:84–90. doi: 10.1111/j.1365-2893.2010.01281.x. [DOI] [PubMed] [Google Scholar]
- Jones SA, Boregowda R, Spratt TE, Hu J. In vitro Epsilon RNA-Dependent Protein Priming Activity of Human Hepatitis B Virus Polymerase. J Virol. 2012;86:5134–5150. doi: 10.1128/JVI.07137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Clark DN, Cao F, Tavis JE, Hu J. Comparative analysis of hepatitis B virus polymerase sequences required for viral RNA binding, RNA packaging, and protein priming. J Virol. 2014;88:1564–1572. doi: 10.1128/JVI.02852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Hu J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging _targets for antiviral intervention. Emerg Microbes Infect. 2013a;2:e56. doi: 10.1038/emi.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Hu J. Protein-primed terminal transferase activity of hepatitis B virus polymerase. J Virol. 2013b;87:2563–2576. doi: 10.1128/JVI.02786-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Murakami E, Delaney W, Furman P, Hu J. Non-competitive Inhibition of Hepatitis B Virus Reverse Transcriptase Protein Priming and DNA synthesis by the Nucleoside Analog Clevudine. Antimicrob Agents Chemother. 2013;57:4181–4189. doi: 10.1128/AAC.00599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SB, Song IH, Kim YM, Noh R, Kang HY, Lee H, Yang HY, Kim AN, Chae HB, Lee SH, Kim HS, Lee TH, Kang YW, Lee ES, Kim SH, Lee BS, Lee HY. Long-term treatment outcomes of clevudine in antiviral-naive patients with chronic hepatitis B. World J Gastroenterol. 2012;18:6943–6950. doi: 10.3748/wjg.v18.i47.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korba BE, Furman PA, Otto MJ. Clevudine: a potent inhibitor of hepatitis B virus in vitro and in vivo. Expert Rev Anti Infect Ther. 2006;4:549–561. doi: 10.1586/14787210.4.4.549. [DOI] [PubMed] [Google Scholar]
- Kwon SY, Park YK, Ahn SH, Cho ES, Choe WH, Lee CH, Kim BK, Ko SY, Choi HS, Park ES, Shin GC, Kim KH. Identification and characterization of clevudine-resistant mutants of hepatitis B virus isolated from chronic hepatitis B patients. Journal of virology. 2010;84:4494–4503. doi: 10.1128/JVI.02066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Notvall L, Beames B. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J Virol. 1995;69:4431–4439. doi: 10.1128/jvi.69.7.4431-4439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Notvall L, Lee H, Beames B. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J Virol. 1997;71:2996–3004. doi: 10.1128/jvi.71.4.2996-3004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Chung YH, Lee K, Byun KS, Paik SW, Han JY, Yoo K, Yoo HW, Lee JH, Yoo BC. A 12-week clevudine therapy showed potent and durable antiviral activity in HBeAg-positive chronic hepatitis B. Hepatology. 2006;43:982–988. doi: 10.1002/hep.21166. [DOI] [PubMed] [Google Scholar]
- Lin L, Hu J. Inhibition of hepadnavirus reverse transcriptase-epsilon RNA interaction by porphyrin compounds. J Virol. 2008;82:2305–2312. doi: 10.1128/JVI.02147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Wan F, Hu J. Functional and structural dynamics of hepadnavirus reverse transcriptase during protein-primed initiation of reverse transcription: effects of metal ions. J Virol. 2008;82:5703–5714. doi: 10.1128/JVI.02760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Arias L, Alvarez M, Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Current opinion in virology. 2014;8C:1–9. doi: 10.1016/j.coviro.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Molnar-Kimber KL, Summers J, Taylor JM, Mason WS. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J Virol. 1983;45:165–172. doi: 10.1128/jvi.45.1.165-172.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C, Bao H, Tolstykh T, Micolochick Steuer HM, Murakami E, Korba B, Furman PA. Evaluation of the in vitro anti-HBV activity of clevudine in combination with other nucleoside/nucleotide inhibitors. Antivir Ther. 2010;15:401–412. doi: 10.3851/IMP1541. [DOI] [PubMed] [Google Scholar]
- Peek SF, Cote PJ, Jacob JR, Toshkov IA, Hornbuckle WE, Baldwin BH, Wells FV, Chu CK, Gerin JL, Tennant BC, Korba BE. Antiviral activity of clevudine [L-FMAU, (1-(2-fluoro-5-methyl-beta, L-arabinofuranosyl) uracil)] against woodchuck hepatitis virus replication and gene expression in chronically infected woodchucks (Marmota monax) Hepatology. 2001;33:254–266. doi: 10.1053/jhep.2001.20899. [DOI] [PubMed] [Google Scholar]
- Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione SJ, Lok AS. Effectiveness of hepatitis B treatment in clinical practice. Gastroenterology. 2012;142:1360–1368. e1361. doi: 10.1053/j.gastro.2012.01.044. [DOI] [PubMed] [Google Scholar]
- Seeger C, Mason WS, Zoulim F. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2977–3029. [Google Scholar]
- Seifer M, Hamatake RK, Colonno RJ, Standring DN. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob Agents Chemother. 1998;42:3200–3208. doi: 10.1128/aac.42.12.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok JI, Lee DK, Lee CH, Park MS, Kim SY, Kim HS, Jo HY, Lee CH, Kim DS. Long-term therapy with clevudine for chronic hepatitis B can be associated with myopathy characterized by depletion of mitochondrial DNA. Hepatology. 2009;49:2080–2086. doi: 10.1002/hep.22959. [DOI] [PubMed] [Google Scholar]
- Skillman AG, Maurer KW, Roe DC, Stauber MJ, Eargle D, Ewing TJ, Muscate A, Davioud-Charvet E, Medaglia MV, Fisher RJ, Arnold E, Gao HQ, Buckheit R, Boyer PL, Hughes SH, Kuntz ID, Kenyon GL. A novel mechanism for inhibition of HIV-1 reverse transcriptase. Bioorg Chem. 2002;30:443–458. doi: 10.1016/s0045-2068(02)00502-3. [DOI] [PubMed] [Google Scholar]
- Stahl M, Beck J, Nassal M. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to epsilon RNA binding. J Virol. 2007a;81:13354–13364. doi: 10.1128/JVI.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Retzlaff M, Nassal M, Beck J. Chaperone activation of the hepadnaviral reverse transcriptase for template RNA binding is established by the Hsp70 and stimulated by the Hsp90 system. Nucleic Acids Res. 2007b;35:6124–6136. doi: 10.1093/nar/gkm628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staschke KA, Colacino JM. Priming of duck hepatitis B virus reverse transcription in vitro: premature termination of primer DNA induced by the 5'-triphosphate of fialuridine. J Virol. 1994;68:8265–8269. doi: 10.1128/jvi.68.12.8265-8269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Tak WY, Yang JM, Kim BI, Baik SK, Cheon GJ, Byun KS, Kim DY, Yoo BC. A randomized, open-label study comparing low-dose clevudine plus adefovir combination therapy with clevudine monotherapy in naive chronic hepatitis B patients. Hepatol Int. 2014;8:375–381. doi: 10.1007/s12072-014-9537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen EJ, Parniak MA, Sarafianos SG. The Hepatitis B Virus Ribonuclease H Is Sensitive to Inhibitors of the Human Immunodeficiency Virus Ribonuclease H and Integrase Enzymes. PLoS Pathog. 2013;9:e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Lomonosova E. The hepatitis B virus ribonuclease H as a drug _target. Antiviral Res. 2015;118:132–138. doi: 10.1016/j.antiviral.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov EP, Obikhod A, Schinazi RF, Gotte M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J Biol Chem. 2008;283:34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Wang GH, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LZ, Kenyon GL, Johnson KA. Novel mechanism of inhibition of HIV-1 reverse transcriptase by a new non-nucleoside analog, KM-1. J Biol Chem. 2004;279:38424–38432. doi: 10.1074/jbc.M406241200. [DOI] [PubMed] [Google Scholar]
- Wang X, Hu J. Distinct requirement for two stages of protein-primed initiation of reverse transcription in hepadnaviruses. J Virol. 2002;76:5857–5865. doi: 10.1128/JVI.76.12.5857-5865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Wen YM, Nassal M. Carbonyl J acid derivatives block protein priming of hepadnaviral P protein and DNA-dependent DNA synthesis activity of hepadnaviral nucleocapsids. J Virol. 2012;86:10079–10092. doi: 10.1128/JVI.00816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Bronsema V, Bartos H, Bosserhoff A, Bartenschlager R, Schaller H. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J Virol. 1994;68:2994–2999. doi: 10.1128/jvi.68.5.2994-2999.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Liu Y, Sui J, Li W. NTCP opens the door for hepatitis B virus infection. Antiviral Res. 2015;121:24–30. doi: 10.1016/j.antiviral.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:1–28. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao GQ, Liu SH, Chou E, Kukhanova M, Chu CK, Cheng YC. Inhibition of Epstein-Barr virus replication by a novel L-nucleoside, 2'-fluoro-5-methyl-beta-L-arabinofuranosyluracil. Biochem Pharmacol. 1996;51:941–947. doi: 10.1016/0006-2952(96)00049-4. [DOI] [PubMed] [Google Scholar]
- Yuen MF, Ahn SH, Lee KS, Um SH, Cho M, Yoon SK, Lee JW, Park NH, Kweon YO, Sohn JH, Lee J, Kim JA, Lai CL, Han KH. Two-year treatment outcome of chronic hepatitis B infection treated with besifovir vs. entecavir: results from a multicentre study. J Hepatol. 2015;62:526–532. doi: 10.1016/j.jhep.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Curtis M, Qi X, Miller MD, Borroto-Esoda K. Anti-hepatitis B virus activity in vitro of combinations of tenofovir with nucleoside/nucleotide analogues. Antiviral chemistry & chemotherapy. 2009;19:165–176. doi: 10.1177/095632020901900404. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. Core protein: A pleiotropic keystone in the HBV lifecycle. Antiviral Res. 2015;121:82–93. doi: 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. e1591–e1592. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68:6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]