Formation of azole-resistant Candida albicans by mutation of sterol 14-demethylase P450
- PMID: 10223930
- PMCID: PMC89127
- DOI: 10.1128/AAC.43.5.1163
Formation of azole-resistant Candida albicans by mutation of sterol 14-demethylase P450
Abstract
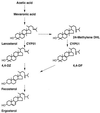
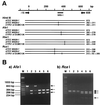
The sterol 14-demethylase P450 (CYP51) of a fluconazole-resistant isolate of Candida albicans, DUMC136, showed reduced susceptibility to this azole but with little change in its catalytic activity. Twelve nucleotide substitutions, resulting in four amino acid changes, were identified in the DUMC136 CYP51 gene in comparison with a reported CYP51 sequence from a wild-type, fluconazole-susceptible C. albicans strain. Seven of these substitutions, including all of those causing amino acid changes, were located within a region covering one of the putative substrate recognition sites of the enzyme (SRS-1). Polymorphisms within this region were observed in several C. albicans isolates, and some were found to be CYP51 heterozygotes. Among the amino acid changes occurring in this region, only an alteration of Y132 was common among these fluconazole-resistant isolates, which suggests the importance of this residue to the fluconazole resistance of the _target enzyme. DUMC136 and another fluconazole-resistant isolate were homozygotes with respect to CYP51, although the typical wild-type, fluconazole-susceptible C. albicans was a CYP51 heterozygote. These findings suggest that part of the fluconazole-resistant phenotype of C. albicans DUMC136 was acquired through a mutation-prone area of CYP51, an area which might promote the formation of fluconazole-resistant CYP51, along with a mechanism(s) which allows the formation of a homozygote of this altered CYP51 in this diploid pathogenic yeast.
Figures

Similar articles
-
Effects of Y132H and F145L substitutions on the activity, azole resistance and spectral properties of Candida albicans sterol 14-demethylase P450 (CYP51): a live example showing the selection of altered P450 through interaction with environmental compounds.J Biochem. 2005 May;137(5):625-32. doi: 10.1093/jb/mvi073. J Biochem. 2005. PMID: 15944416
-
Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents.Antimicrob Agents Chemother. 1998 Feb;42(2):241-53. doi: 10.1128/AAC.42.2.241. Antimicrob Agents Chemother. 1998. PMID: 9527767 Free PMC article.
-
Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans.Microbiology (Reading). 1999 Oct;145 ( Pt 10):2701-2713. doi: 10.1099/00221287-145-10-2701. Microbiology (Reading). 1999. PMID: 10537192
-
Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature.Diagn Microbiol Infect Dis. 2010 Apr;66(4):373-84. doi: 10.1016/j.diagmicrobio.2009.11.006. Diagn Microbiol Infect Dis. 2010. PMID: 20226328 Review.
-
[Azole resistance in Candida spp].Nihon Ishinkin Gakkai Zasshi. 2003;44(2):87-92. doi: 10.3314/jjmm.44.87. Nihon Ishinkin Gakkai Zasshi. 2003. PMID: 12748589 Review. Japanese.
Cited by
-
The development of fluconazole resistance in Candida albicans - an example of microevolution of a fungal pathogen.J Microbiol. 2016 Mar;54(3):192-201. doi: 10.1007/s12275-016-5628-4. Epub 2016 Feb 27. J Microbiol. 2016. PMID: 26920879 Review.
-
Possible inhibitory molecular mechanism of farnesol on the development of fluconazole resistance in Candida albicans biofilm.Antimicrob Agents Chemother. 2012 Feb;56(2):770-5. doi: 10.1128/AAC.05290-11. Epub 2011 Nov 21. Antimicrob Agents Chemother. 2012. PMID: 22106223 Free PMC article.
-
Species distribution and antifungal susceptibility patterns of Candida spp. bloodstream isolates from a Brazilian tertiary care hospital.Mycopathologia. 2007 Mar;163(3):145-51. doi: 10.1007/s11046-007-0094-5. Epub 2007 Mar 1. Mycopathologia. 2007. PMID: 17334813
-
Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients.Antimicrob Agents Chemother. 2001 Oct;45(10):2676-84. doi: 10.1128/AAC.45.10.2676-2684.2001. Antimicrob Agents Chemother. 2001. PMID: 11557454 Free PMC article.
-
The Impact of Gene Dosage and Heterozygosity on The Diploid Pathobiont Candida albicans.J Fungi (Basel). 2019 Dec 27;6(1):10. doi: 10.3390/jof6010010. J Fungi (Basel). 2019. PMID: 31892130 Free PMC article. Review.
References
-
- Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y, Kurosawa N, Horiuchi T, Yoshida Y. Sterol 14-demethylase P450 (P45014DM) is one of the most ancient and conserved P450 species. J Biochem. 1996;119:926–933. - PubMed
-
- Aoyama Y, Yoshida Y, Sato R. Yeast cytochrome P-450 catalyzing lanosterol 14α-demethylation. J Biol Chem. 1984;259:1661–1666. - PubMed
-
- Barrett-Bee K J, Lane A C, Turner R W. The mode of antifungal action of tolnaftate. J Med Vet Mycol. 1986;24:155–160. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

