Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage
- PMID: 10570149
- PMCID: PMC24141
- DOI: 10.1073/pnas.96.24.13777
Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage
Abstract
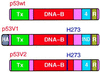
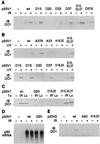
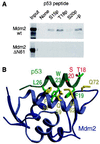
Stabilization of p53 in response to DNA damage is caused by its dissociation from Mdm2, a protein that _targets p53 for degradation in the proteasome. Dissociation of p53 from Mdm2 could be caused by DNA damage-induced p53 posttranslational modifications. The ATM and ATR kinases, whose activation in response to ionizing radiation (IR) and UV light, respectively, is required for p53 stabilization, directly phosphorylate p53 on Ser-15. However, phosphorylation of Ser-15 is critical for the apoptotic activity of p53 and not for p53 stabilization. Thus, whether any p53 modifications, and which, underlie disruption of the p53-Mdm2 complex after DNA damage remains to be determined. We analyzed the IR- and UV light-induced stabilization of p53 proteins with substitutions of Ser known to be posttranslationally modified after DNA damage. Substitution of Ser-20 was sufficient to abrogate p53 stabilization in response to both IR and UV light. Furthermore, both IR and UV light induced phosphorylation of p53 on Ser-20, which involved the majority of nuclear p53 protein and weakened the interaction of p53 with Mdm2 in vitro. ATM and ATR cannot phosphorylate p53 on Ser-20. We therefore propose that ATM and ATR activate an, as yet unidentified, kinase that stabilizes p53 by phosphorylating it on Ser-20.
Figures


Similar articles
-
Phosphorylation of murine double minute clone 2 (MDM2) protein at serine-267 by protein kinase CK2 in vitro and in cultured cells.Biochem J. 2001 Apr 15;355(Pt 2):347-56. doi: 10.1042/0264-6021:3550347. Biochem J. 2001. PMID: 11284721 Free PMC article.
-
ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage.Genes Dev. 2001 May 1;15(9):1067-77. doi: 10.1101/gad.886901. Genes Dev. 2001. PMID: 11331603 Free PMC article.
-
Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage.Proc Natl Acad Sci U S A. 1999 Dec 21;96(26):14973-7. doi: 10.1073/pnas.96.26.14973. Proc Natl Acad Sci U S A. 1999. PMID: 10611322 Free PMC article.
-
Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress.Neoplasia. 2000 May-Jun;2(3):208-25. doi: 10.1038/sj.neo.7900073. Neoplasia. 2000. PMID: 10935507 Free PMC article. Review.
-
A closer view of an oncoprotein-tumor suppressor interaction.Chem Biol. 1997 Nov;4(11):791-4. doi: 10.1016/s1074-5521(97)90112-5. Chem Biol. 1997. PMID: 9384532 Review.
Cited by
-
Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype.Front Cell Dev Biol. 2021 Mar 29;9:645593. doi: 10.3389/fcell.2021.645593. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33855023 Free PMC article. Review.
-
Tumor suppressor p53: Biology, signaling pathways, and therapeutic _targeting.Biochim Biophys Acta Rev Cancer. 2021 Aug;1876(1):188556. doi: 10.1016/j.bbcan.2021.188556. Epub 2021 Apr 29. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33932560 Free PMC article. Review.
-
Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation.EMBO J. 2004 Apr 7;23(7):1547-56. doi: 10.1038/sj.emboj.7600145. Epub 2004 Mar 18. EMBO J. 2004. PMID: 15029243 Free PMC article.
-
Temperature-sensitive ovarian carcinoma cell line (OvBH-1).Jpn J Cancer Res. 2002 Sep;93(9):976-85. doi: 10.1111/j.1349-7006.2002.tb02473.x. Jpn J Cancer Res. 2002. PMID: 12359050 Free PMC article.
-
Posttranscriptional regulation of p53 and its _targets by RNA-binding proteins.Curr Mol Med. 2008 Dec;8(8):845-9. doi: 10.2174/156652408786733748. Curr Mol Med. 2008. PMID: 19075680 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

