Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53
- PMID: 10673500
- PMCID: PMC316357
Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53
Abstract
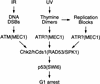
Chk2/hcds1, the human homolog of the Saccharomyces cerevisiae RAD53/SPK1 and Schizosaccharomyces pombe cds1 DNA damage checkpoint genes, encodes a protein kinase that is post-translationally modified after DNA damage. Like its yeast homologs, the Chk2/hCds1 protein phosphorylates Cdc25C in vitro, suggesting that it arrests cells in G(2) in response to DNA damage. We expressed Chk2/hCds1 in human cells and analyzed their cell cycle profile. Wild-type, but not catalytically inactive, Chk2/hCds1 led to G(1) arrest after DNA damage. The arrest was inhibited by cotransfection of a dominant-negative p53 mutant, indicating that Chk2/hCds1 acted upstream of p53. In vitro, Chk2/hCds1 phosphorylated p53 on Ser-20 and dissociated preformed complexes of p53 with Mdm2, a protein that _targets p53 for degradation. In vivo, ectopic expression of wild-type Chk2/hCds1 led to increased p53 stabilization after DNA damage, whereas expression of a dominant-negative Chk2/hCds1 mutant abrogated both phosphorylation of p53 on Ser-20 and p53 stabilization. Thus, in response to DNA damage, Chk2/hCds1 stabilizes the p53 tumor suppressor protein leading to cell cycle arrest in G(1).
Figures






Similar articles
-
Role of human Cds1 (Chk2) kinase in DNA damage checkpoint and its regulation by p53.J Biol Chem. 1999 Oct 29;274(44):31463-7. doi: 10.1074/jbc.274.44.31463. J Biol Chem. 1999. PMID: 10531348
-
The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites.Genes Dev. 2000 Feb 1;14(3):289-300. Genes Dev. 2000. PMID: 10673501 Free PMC article.
-
DNA damage-induced activation of p53 by the checkpoint kinase Chk2.Science. 2000 Mar 10;287(5459):1824-7. doi: 10.1126/science.287.5459.1824. Science. 2000. PMID: 10710310
-
How to activate p53.Curr Biol. 2000 Apr 20;10(8):R315-7. doi: 10.1016/s0960-9822(00)00439-5. Curr Biol. 2000. PMID: 10801407 Review.
-
Checking in on Cds1 (Chk2): A checkpoint kinase and tumor suppressor.Bioessays. 2002 Jun;24(6):502-11. doi: 10.1002/bies.10101. Bioessays. 2002. PMID: 12111733 Review.
Cited by
-
SMG7 is a critical regulator of p53 stability and function in DNA damage stress response.Cell Discov. 2016 Jan 19;2:15042. doi: 10.1038/celldisc.2015.42. eCollection 2016. Cell Discov. 2016. PMID: 27462439 Free PMC article.
-
Mechanism-based screen establishes signalling framework for DNA damage-associated G1 checkpoint response.PLoS One. 2012;7(2):e31627. doi: 10.1371/journal.pone.0031627. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22384045 Free PMC article.
-
Inhibition of the ATM/Chk2 axis promotes cGAS/STING signaling in ARID1A-deficient tumors.J Clin Invest. 2020 Nov 2;130(11):5951-5966. doi: 10.1172/JCI130445. J Clin Invest. 2020. PMID: 33016929 Free PMC article.
-
Regulation of the Human Papillomavirus Life Cycle by DNA Damage Repair Pathways and Epigenetic Factors.Viruses. 2020 Jul 10;12(7):744. doi: 10.3390/v12070744. Viruses. 2020. PMID: 32664381 Free PMC article. Review.
-
The co-repressor SMRT delays DNA damage-induced caspase activation by repressing pro-apoptotic genes and modulating the dynamics of checkpoint kinase 2 activation.PLoS One. 2013 May 17;8(5):e59986. doi: 10.1371/journal.pone.0059986. Print 2013. PLoS One. 2013. PMID: 23690919 Free PMC article.
References
-
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
-
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, et al. Heterozygous germline hChk2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
