Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria
- PMID: 10684930
- PMCID: PMC111038
- DOI: 10.1093/nar/28.6.1355
Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria
Erratum in
-
Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria.Nucleic Acids Res. 2015 Apr 20;43(7):3870-1. doi: 10.1093/nar/gkv264. Epub 2015 Mar 23. Nucleic Acids Res. 2015. PMID: 25800745 Free PMC article. No abstract available.
Abstract
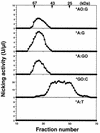
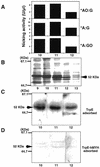
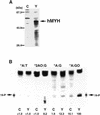
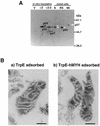
An enzyme activity introducing an alkali-labile site at 2-hydroxyadenine (2-OH-A) in double-stranded oligonucleotides was detected in nuclear extracts of Jurkat cells. This activity co-eluted with activities toward adenine paired with guanine and 8-oxo-7,8-dihydroguanine (8-oxoG) as a single peak corresponding to a 55 kDa molecular mass on gel filtration chromatography. Further co-purification was then done. Western blotting revealed that these activities also co-purified with a 52 kDa polypeptide which reacted with antibodies against human MYH (anti-hMYH). Recombinant hMYH has essentially similar activities to the partially purified enzyme. Thus, hMYH is likely to possess both adenine and 2-OH-A DNA glycosylase activities. In nuclear extracts from Jurkat cells, a 52 kDa polypeptide was detected with a small amount of 53 kDa polypeptide, while in mitochondrial extracts a 57 kDa polypeptide was detected using anti-hMYH. With amplification of the 5'-regions of the hMYH cDNA, 10 forms of hMYH transcripts were identified and subgrouped into three types, each with a unique 5' sequence. These hMYH transcripts are likely to encode multiple authentic hMYH polypeptides including the 52, 53 and 57 kDa polypeptides detected in Jurkat cells.
Figures




Similar articles
-
Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria.Nucleic Acids Res. 2000 Sep 1;28(17):3206-15. doi: 10.1093/nar/28.17.3206. Nucleic Acids Res. 2000. PMID: 10954587 Free PMC article.
-
Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase.Nucleic Acids Res. 1999 Sep 15;27(18):3638-44. doi: 10.1093/nar/27.18.3638. Nucleic Acids Res. 1999. PMID: 10471731 Free PMC article.
-
Differential DNA recognition and glycosylase activity of the native human MutY homolog (hMYH) and recombinant hMYH expressed in bacteria.Nucleic Acids Res. 2001 Jun 15;29(12):2666-74. doi: 10.1093/nar/29.12.2666. Nucleic Acids Res. 2001. PMID: 11410677 Free PMC article.
-
Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage.Prog Nucleic Acid Res Mol Biol. 2001;68:75-94. doi: 10.1016/s0079-6603(01)68091-7. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554314 Review.
-
Human MutY: gene structure, protein functions and interactions, and role in carcinogenesis.Cell Mol Life Sci. 2003 Oct;60(10):2064-83. doi: 10.1007/s00018-003-3053-4. Cell Mol Life Sci. 2003. PMID: 14618256 Free PMC article. Review.
Cited by
-
The maintenance of mitochondrial DNA integrity--critical analysis and update.Cold Spring Harb Perspect Biol. 2013 May 1;5(5):a012641. doi: 10.1101/cshperspect.a012641. Cold Spring Harb Perspect Biol. 2013. PMID: 23637283 Free PMC article. Review.
-
Dynamic features of human mitochondrial DNA maintenance and transcription.Front Cell Dev Biol. 2022 Sep 7;10:984245. doi: 10.3389/fcell.2022.984245. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36158192 Free PMC article. Review.
-
Mitochondrial Oxidative Stress, Mitochondrial DNA Damage and Their Role in Age-Related Vascular Dysfunction.Int J Mol Sci. 2015 Jul 13;16(7):15918-53. doi: 10.3390/ijms160715918. Int J Mol Sci. 2015. PMID: 26184181 Free PMC article. Review.
-
Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria.Nucleic Acids Res. 2000 Sep 1;28(17):3206-15. doi: 10.1093/nar/28.17.3206. Nucleic Acids Res. 2000. PMID: 10954587 Free PMC article.
-
Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen.Nucleic Acids Res. 2001 Jun 1;29(11):2349-60. doi: 10.1093/nar/29.11.2349. Nucleic Acids Res. 2001. PMID: 11376153 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

