An aminoacylation-dependent nuclear tRNA export pathway in yeast
- PMID: 10766739
- PMCID: PMC316491
An aminoacylation-dependent nuclear tRNA export pathway in yeast
Abstract
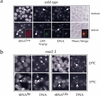
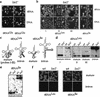
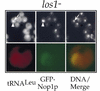
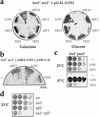
Yeast Los1p, the homolog of human exportin-t, mediates nuclear export of tRNA. Using fluorescence in situ hybridization, we could show that the export of some intronless tRNA species is not detectably affected by the disruption of LOS1. To find other factors that facilitate tRNA export, we performed a suppressor screen of a synthetically lethal los1 mutant and identified the essential translation elongation factor eEF-1A. Mutations in eEF-1A impaired nuclear export of all tRNAs tested, which included both spliced and intronless species. An even stronger defect in nuclear exit of tRNA was observed under conditions that inhibited tRNA aminoacylation. In all cases, inhibition of tRNA export led to nucleolar accumulation of mature tRNAs. Our data show that tRNA aminoacylation and eEF-1A are required for efficient nuclear tRNA export in yeast and suggest coordination between the protein translation and the nuclear tRNA processing and transport machineries.
Figures



Similar articles
-
The nuclear tRNA aminoacylation-dependent pathway may be the principal route used to export tRNA from the nucleus in Saccharomyces cerevisiae.Biochem J. 2004 Mar 15;378(Pt 3):809-16. doi: 10.1042/BJ20031306. Biochem J. 2004. PMID: 14640976 Free PMC article.
-
Cex1p is a novel cytoplasmic component of the Saccharomyces cerevisiae nuclear tRNA export machinery.EMBO J. 2007 Jan 24;26(2):288-300. doi: 10.1038/sj.emboj.7601493. Epub 2007 Jan 4. EMBO J. 2007. PMID: 17203074 Free PMC article.
-
Review: transport of tRNA out of the nucleus-direct channeling to the ribosome?J Struct Biol. 2000 Apr;129(2-3):288-94. doi: 10.1006/jsbi.2000.4226. J Struct Biol. 2000. PMID: 10806079 Review.
-
Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export.Mol Biol Cell. 2001 May;12(5):1381-92. doi: 10.1091/mbc.12.5.1381. Mol Biol Cell. 2001. PMID: 11359929 Free PMC article.
-
The ins and outs of nuclear re-export of retrogradely transported tRNAs in Saccharomyces cerevisiae.Nucleus. 2010 May-Jun;1(3):224-30. doi: 10.4161/nucl.1.3.11250. Epub 2010 Jan 13. Nucleus. 2010. PMID: 21327067 Free PMC article. Review.
Cited by
-
The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability.RNA. 2006 Apr;12(4):644-54. doi: 10.1261/rna.2307206. RNA. 2006. PMID: 16581807 Free PMC article.
-
The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p.EMBO J. 2001 Dec 3;20(23):6889-98. doi: 10.1093/emboj/20.23.6889. EMBO J. 2001. PMID: 11726524 Free PMC article.
-
Cyclophilin A peptidyl-prolyl isomerase activity promotes ZPR1 nuclear export.Mol Cell Biol. 2002 Oct;22(20):6993-7003. doi: 10.1128/MCB.22.20.6993-7003.2002. Mol Cell Biol. 2002. PMID: 12242280 Free PMC article.
-
Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes.Nucleic Acids Res. 2006;34(14):3968-79. doi: 10.1093/nar/gkl560. Epub 2006 Aug 12. Nucleic Acids Res. 2006. PMID: 16914447 Free PMC article.
-
tRNA trafficking along the TOR pathway.Cell Cycle. 2010 Aug 15;9(16):3146-7. doi: 10.4161/cc.9.16.12781. Epub 2010 Aug 15. Cell Cycle. 2010. PMID: 20814229 Free PMC article. No abstract available.
References
-
- Adam SA. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. - PubMed
-
- Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, Abelson J. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265:16216–16220. - PubMed
-
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. - PubMed
-
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998a;8:305–314. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
