Single-molecule protein folding: diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2
- PMID: 10792044
- PMCID: PMC25802
- DOI: 10.1073/pnas.090104997
Single-molecule protein folding: diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2
Abstract
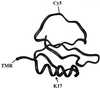
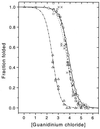
We report single-molecule folding studies of a small, single-domain protein, chymotrypsin inhibitor 2 (CI2). CI2 is an excellent model system for protein folding studies and has been extensively studied, both experimentally (at the ensemble level) and theoretically. Conformationally assisted ligation methodology was used to synthesize the proteins and site-specifically label them with donor and acceptor dyes. Folded and denatured subpopulations were observed by fluorescence resonance energy transfer (FRET) measurements on freely diffusing single protein molecules. Properties of these subpopulations were directly monitored as a function of guanidinium chloride concentration. It is shown that new information about different aspects of the protein folding reaction can be extracted from such subpopulation properties. Shifts in the mean transfer efficiencies are discussed, FRET efficiency distributions are translated into potentials, and denaturation curves are directly plotted from the areas of the FRET peaks. Changes in stability caused by mutation also are measured by comparing pseudo wild-type CI2 with a destabilized mutant (K17G). Current limitations and future possibilities and prospects for single-pair FRET protein folding investigations are discussed.
Figures


Similar articles
-
Complementation of peptide fragments of the single domain protein chymotrypsin inhibitor 2.J Mol Biol. 1997 Oct 17;273(1):317-29. doi: 10.1006/jmbi.1997.1303. J Mol Biol. 1997. PMID: 9367764
-
The structure of the transition state for folding of chymotrypsin inhibitor 2 analysed by protein engineering methods: evidence for a nucleation-condensation mechanism for protein folding.J Mol Biol. 1995 Nov 24;254(2):260-88. doi: 10.1006/jmbi.1995.0616. J Mol Biol. 1995. PMID: 7490748
-
The rate of isomerisation of peptidyl-proline bonds as a probe for interactions in the physiological denatured state of chymotrypsin inhibitor 2.J Mol Biol. 1997 Jun 20;269(4):611-22. doi: 10.1006/jmbi.1997.1043. J Mol Biol. 1997. PMID: 9217264
-
The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements.Chemphyschem. 2005 May;6(5):858-70. doi: 10.1002/cphc.200400617. Chemphyschem. 2005. PMID: 15884068 Review.
-
Association of complementary fragments and the elucidation of protein folding pathways.Protein Eng. 1996 Oct;9(10):843-7. doi: 10.1093/protein/9.10.843. Protein Eng. 1996. PMID: 8931123 Review.
Cited by
-
Observation of persistent α-helical content and discrete types of backbone disorder during a molten globule to ordered peptide transition via deep-UV resonance Raman spectroscopy.J Raman Spectrosc. 2013 Jul;44(7):957-962. doi: 10.1002/jrs.4316. Epub 2013 Jun 1. J Raman Spectrosc. 2013. PMID: 27795611 Free PMC article.
-
Dynamics of synaptic SfiI-DNA complex: single-molecule fluorescence analysis.Biophys J. 2007 May 1;92(9):3241-50. doi: 10.1529/biophysj.106.095778. Epub 2007 Feb 2. Biophys J. 2007. PMID: 17277188 Free PMC article.
-
A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures.Proc Natl Acad Sci U S A. 2007 Feb 20;104(8):2649-54. doi: 10.1073/pnas.0611503104. Epub 2007 Feb 13. Proc Natl Acad Sci U S A. 2007. PMID: 17299036 Free PMC article.
-
Choosing the Probe for Single-Molecule Fluorescence Microscopy.Int J Mol Sci. 2022 Nov 29;23(23):14949. doi: 10.3390/ijms232314949. Int J Mol Sci. 2022. PMID: 36499276 Free PMC article. Review.
-
Distinguishing between protein dynamics and dye photophysics in single-molecule FRET experiments.Biophys J. 2010 Feb 17;98(4):696-706. doi: 10.1016/j.bpj.2009.12.4322. Biophys J. 2010. PMID: 20159166 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

