Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5'-nuclease assays
- PMID: 11055900
- PMCID: PMC92356
- DOI: 10.1128/AEM.66.11.4605-4614.2000
Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5'-nuclease assays
Abstract
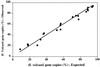
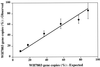
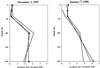
Few techniques are currently available for quantifying specific prokaryotic taxa in environmental samples. Quantification of specific genotypes has relied mainly on oligonucleotide hybridization to extracted rRNA or intact rRNA in whole cells. However, low abundance and cellular rRNA content limit the application of these techniques in aquatic environments. In this study, we applied a newly developed quantitative PCR assay (5'-nuclease assay, also known as TaqMan) to quantify specific small-subunit (SSU) rRNA genes (rDNAs) from uncultivated planktonic prokaryotes in Monterey Bay. Primer and probe combinations for quantification of SSU rDNAs at the domain and group levels were developed and tested for specificity and quantitative reliability. We examined the spatial and temporal variations of SSU rDNAs from Synechococcus plus Prochlorococcus and marine Archaea and compared the results of the quantitative PCR assays to those obtained by alternative methods. The 5'-nuclease assays reliably quantified rDNAs over at least 4 orders of magnitude and accurately measured the proportions of genes in artificial mixtures. The spatial and temporal distributions of planktonic microbial groups measured by the 5'-nuclease assays were similar to the distributions estimated by quantitative oligonucleotide probe hybridization, whole-cell hybridization assays, and flow cytometry.
Figures


Similar articles
-
Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes.Appl Environ Microbiol. 2000 Nov;66(11):5066-72. doi: 10.1128/AEM.66.11.5066-5072.2000. Appl Environ Microbiol. 2000. PMID: 11055964 Free PMC article.
-
Comparison of real-time PCR with SYBR Green I or 5'-nuclease assays and dot-blot hybridization with rDNA-_targeted oligonucleotide probes in quantification of selected faecal bacteria.Microbiology (Reading). 2003 Jan;149(Pt 1):269-77. doi: 10.1099/mic.0.25975-0. Microbiology (Reading). 2003. PMID: 12576600
-
Development and application of a real-time PCR approach for quantification of uncultured bacteria in the central Baltic Sea.Appl Environ Microbiol. 2004 Aug;70(8):4971-9. doi: 10.1128/AEM.70.8.4971-4979.2004. Appl Environ Microbiol. 2004. PMID: 15294837 Free PMC article.
-
Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques.Nat Rev Microbiol. 2008 May;6(5):339-48. doi: 10.1038/nrmicro1888. Nat Rev Microbiol. 2008. PMID: 18414500 Review.
-
Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis.FEMS Microbiol Rev. 1997 Nov;21(3):213-29. doi: 10.1111/j.1574-6976.1997.tb00351.x. FEMS Microbiol Rev. 1997. PMID: 9451814 Review.
Cited by
-
Prevalence and Fate of Carbapenemase Genes in a Wastewater Treatment Plant in Northern China.PLoS One. 2016 May 26;11(5):e0156383. doi: 10.1371/journal.pone.0156383. eCollection 2016. PLoS One. 2016. PMID: 27227329 Free PMC article.
-
Community structure, abundance, and activity of methanotrophs in the Zoige wetland of the Tibetan Plateau.Microb Ecol. 2012 May;63(4):835-43. doi: 10.1007/s00248-011-9981-x. Epub 2011 Dec 10. Microb Ecol. 2012. PMID: 22159497
-
Oil field microorganisms cause highly localized corrosion on chemically inhibited carbon steel.Microb Biotechnol. 2021 Jan;14(1):171-185. doi: 10.1111/1751-7915.13644. Epub 2020 Sep 17. Microb Biotechnol. 2021. PMID: 32940951 Free PMC article.
-
Homogeneity and synchronous dynamics of microbial communities in particulate biofilms: from major populations to minor groups.Microbes Environ. 2012;27(2):142-8. doi: 10.1264/jsme2.me11264. Microbes Environ. 2012. PMID: 22791046 Free PMC article.
-
Comparative evaluation of establishing a human gut microbial community within rodent models.Gut Microbes. 2012 May-Jun;3(3):234-49. doi: 10.4161/gmic.19934. Epub 2012 May 1. Gut Microbes. 2012. PMID: 22572831 Free PMC article.
References
-
- Ausubel F A, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1988.
-
- Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorte, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. Environ. Microbiol., in press. - PubMed
-
- Buck K R, Chavez F P, Campbell L. Basin-wide distributions of living carbon components and the inverted trophic pyramid of the central gyre of the North Atlantic Ocean, summer 1993. Aquat Microb Ecol. 1996;10:283–298.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

