Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting
- PMID: 11080168
- PMCID: PMC305813
- DOI: 10.1093/emboj/19.22.6230
Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting
Abstract
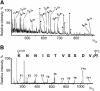
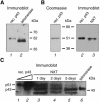
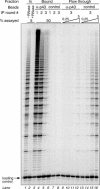
Telomerase is the ribonucleoprotein enzyme responsible for the replication of chromosome ends in most eukaryotes. In the ciliate Euplotes aediculatus, the protein p43 biochemically co-purifies with active telomerase and appears to be stoichiometric with both the RNA and the catalytic protein subunit of this telomerase complex. Here we describe cloning of the gene for p43 and present evidence that it is an authentic component of the telomerase holoenzyme. Comparison of the nucleotide sequence of the cloned gene with peptide sequences of the protein suggests that production of full-length p43 relies on a programmed ribosomal frameshift, an extremely rare translational mechanism. Anti-p43 antibodies immunodeplete telomerase RNA and telomerase activity from E.aediculatus nuclear extracts, indicating that the vast majority of mature telomerase complexes in the cell are associated with p43. The sequence of p43 reveals similarity to the La autoantigen, an RNA-binding protein involved in maturation of RNA polymerase III transcripts, and recombinant p43 binds telomerase RNA in vitro. By analogy to other La proteins, p43 may function in chaperoning the assembly and/or facilitating nuclear retention of telomerase.
Figures

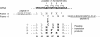



Similar articles
-
The Euplotes La motif protein p43 has properties of a telomerase-specific subunit.Biochemistry. 2003 May 20;42(19):5736-47. doi: 10.1021/bi034121y. Biochemistry. 2003. PMID: 12741831
-
The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro.RNA. 2004 Jul;10(7):1108-18. doi: 10.1261/rna.7400704. RNA. 2004. PMID: 15208446 Free PMC article.
-
The telomerase-associated protein p43 is involved in anchoring telomerase in the nucleus.J Cell Sci. 2003 May 1;116(Pt 9):1757-61. doi: 10.1242/jcs.00351. J Cell Sci. 2003. PMID: 12665556
-
Telomerase and the chromosome end replication problem.Ciba Found Symp. 1997;211:20-8; discussion 28-34. doi: 10.1002/9780470515433.ch3. Ciba Found Symp. 1997. PMID: 9524749 Review.
-
Telomerase is a true reverse transcriptase. A review.Biochemistry (Mosc). 1997 Nov;62(11):1202-5. Biochemistry (Mosc). 1997. PMID: 9467843 Review.
Cited by
-
Positive and negative regulation of Tetrahymena telomerase holoenzyme.Mol Cell Biol. 2007 Mar;27(6):2074-83. doi: 10.1128/MCB.02105-06. Epub 2007 Jan 12. Mol Cell Biol. 2007. PMID: 17220281 Free PMC article.
-
Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.Mol Cell. 2022 May 5;82(9):1724-1736.e7. doi: 10.1016/j.molcel.2022.03.001. Epub 2022 Mar 22. Mol Cell. 2022. PMID: 35320752 Free PMC article.
-
The La antigen associates with the human telomerase ribonucleoprotein and influences telomere length in vivo.RNA. 2001 Aug;7(8):1068-75. doi: 10.1017/s1355838201010159. RNA. 2001. PMID: 11497426 Free PMC article.
-
A comprehensive analysis of the La-motif protein superfamily.RNA. 2009 May;15(5):750-64. doi: 10.1261/rna.1478709. Epub 2009 Mar 19. RNA. 2009. PMID: 19299548 Free PMC article.
-
LARP3, LARP7, and MePCE are involved in the early stage of human telomerase RNA biogenesis.Nat Commun. 2024 Jul 15;15(1):5955. doi: 10.1038/s41467-024-50422-w. Nat Commun. 2024. PMID: 39009594 Free PMC article.
References
-
- Bandziulis R.J., Swanson,M.S. and Dreyfuss,G. (1989) RNA-binding proteins as developmental regulators. Genes Dev., 3, 431–437. - PubMed
-
- Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. - PubMed
-
- Blasco M.A, Lee,H.-W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.E. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

