The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins
- PMID: 11266464
- PMCID: PMC2195777
- DOI: 10.1083/jcb.152.4.729
The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins
Abstract
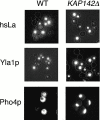
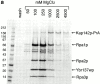
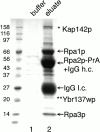
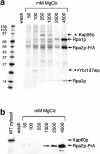
We have identified a novel pathway for protein import into the nucleus. Although the product of Saccharomyces cerevisiae gene MSN5 was previously shown to function as a karyopherin (Kap) for nuclear export of various proteins, we discovered a nuclear import pathway mediated by Msn5p (also referred to as Kap142p). We have purified from yeast cytosol a complex containing Kap142p and the trimeric replication protein A (RPA), which is required for multiple aspects of DNA metabolism, including DNA replication, DNA repair, and recombination. In wild-type cells, RPA was localized primarily to the nucleus but, in a KAP142 deletion strain, RPA was mislocalized to the cytoplasm and the strain was highly sensitive to bleomycin (BLM). BLM causes DNA double-strand breaks and, in S. cerevisiae, the DNA damage is repaired predominantly by RPA-dependent homologous recombination. Therefore, our results indicate that in wild-type cells a critical portion of RPA was imported into the nucleus by Kap142p. Like several other import-related Kap-substrate complexes, the endogenous RPA-Kap142p complex was dissociated by RanGTP, but not by RanGDP. All three RPA genes are essential for viability, whereas KAP142 is not. Perhaps explaining this disparity, we observed an interaction between RPA and Kap95p in a strain lacking Kap142p. This interaction could provide a mechanism for import of RPA into the nucleus and cell viability in the absence of Kap142p. Together with published results (Kaffman, A., N.M. Rank, E.M. O'Neill, L.S. Huang, and E.K. O'Shea. 1998. Nature. 396:482-486; Blondel, M., P.M. Alepuz, L.S. Huang, S. Shaham, G. Ammerer, and M. Peter. 1999. Genes Dev. 13:2284-2300; DeVit, M.J., and M. Johnston. 1999. Curr. Biol. 9:1231-1241; Mahanty, S.K., Y. Wang, F.W. Farley, and E.A. Elion. 1999. Cell. 98:501-512) our data indicate that the karyopherin Kap142p is able to mediate nuclear import of one set of proteins and nuclear export of a different set of proteins.
Figures







Similar articles
-
Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins.J Cell Biol. 2002 Jan 7;156(1):53-64. doi: 10.1083/jcb.200110082. Epub 2002 Jan 3. J Cell Biol. 2002. PMID: 11777942 Free PMC article.
-
The Saccharomyces cerevisiae RanGTP-binding protein msn5p is involved in different signal transduction pathways.Genetics. 1999 Nov;153(3):1219-31. doi: 10.1093/genetics/153.3.1219. Genetics. 1999. PMID: 10545454 Free PMC article.
-
The karyopherin Kap95 and the C-termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae.DNA Cell Biol. 2011 Sep;30(9):641-51. doi: 10.1089/dna.2010.1071. Epub 2011 Feb 20. DNA Cell Biol. 2011. PMID: 21332387 Free PMC article.
-
Nuclear import of histones.Biochem Soc Trans. 2020 Dec 18;48(6):2753-2767. doi: 10.1042/BST20200572. Biochem Soc Trans. 2020. PMID: 33300986 Free PMC article. Review.
-
Distinct nuclear import and export pathways mediated by members of the karyopherin beta family.J Cell Biochem. 1998 Aug 1;70(2):231-9. J Cell Biochem. 1998. PMID: 9671229 Review.
Cited by
-
Essential global role of CDC14 in DNA synthesis revealed by chromosome underreplication unrecognized by checkpoints in cdc14 mutants.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14466-71. doi: 10.1073/pnas.0900190106. Epub 2009 Aug 7. Proc Natl Acad Sci U S A. 2009. PMID: 19666479 Free PMC article.
-
Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling.Mol Cell Biol. 2003 May;23(9):3126-40. doi: 10.1128/MCB.23.9.3126-3140.2003. Mol Cell Biol. 2003. PMID: 12697814 Free PMC article.
-
RanGTPase: A Key Regulator of Nucleocytoplasmic Trafficking.Mol Cell Pharmacol. 2009;1(3):148-156. doi: 10.4255/mcpharmacol.09.19. Mol Cell Pharmacol. 2009. PMID: 20300488 Free PMC article.
-
A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomycescerevisiae.Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5412-7. doi: 10.1073/pnas.082682699. Proc Natl Acad Sci U S A. 2002. PMID: 11959996 Free PMC article.
-
Engineered SUMO/protease system identifies Pdr6 as a bidirectional nuclear transport receptor.J Cell Biol. 2019 Jun 3;218(6):2006-2020. doi: 10.1083/jcb.201812091. Epub 2019 Apr 25. J Cell Biol. 2019. PMID: 31023724 Free PMC article.
References
-
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup157pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. - PMC - PubMed
-
- Aitchison J.D., Blobel G., Rout M.P. Kap104pa karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

