SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome
- PMID: 11331595
- PMCID: PMC125440
- DOI: 10.1093/emboj/20.9.2304
SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome
Abstract
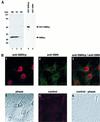
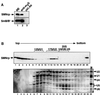
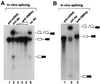
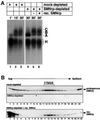
SMNrp, also termed SPF30, has recently been identified in spliceosomes assembled in vitro. We have functionally characterized this protein and show that it is an essential splicing factor. We show that SMNrp is a 17S U2 snRNP-associated protein that appears in the pre-spliceosome (complex A) and the mature spliceosome (complex B) during splicing. Immunodepletion of SMNrp from nuclear extract inhibits the first step of pre-mRNA splicing by preventing the formation of complex B. Re-addition of recombinant SMNrp to immunodepleted extract reconstitutes both spliceosome formation and splicing. Mutations in two domains of SMNrp, although similarly deleterious for splicing, differed in their consequences on U2 snRNP binding, suggesting that SMNrp may also engage in interactions with splicing factors other than the U2 snRNP. In agreement with this, we present evidence for an additional interaction between SMNrp and the [U4/U6.U5] tri-snRNP. A candidate that may mediate this interaction, namely the U4/U6-90 kDa protein, has been identified. We suggest that SMNrp, as a U2 snRNP-associated protein, facilitates the recruitment of the [U4/U6.U5] tri-snRNP to the pre-spliceosome.
Figures







Similar articles
-
SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome.J Biol Chem. 2001 Aug 17;276(33):31142-50. doi: 10.1074/jbc.M103620200. Epub 2001 Apr 30. J Biol Chem. 2001. PMID: 11331295
-
The splicing factor Prp17 interacts with the U2, U5 and U6 snRNPs and associates with the spliceosome pre- and post-catalysis.Biochem J. 2008 Dec 15;416(3):365-74. doi: 10.1042/BJ20081195. Biochem J. 2008. PMID: 18691155
-
Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing.EMBO J. 2002 Mar 1;21(5):1148-57. doi: 10.1093/emboj/21.5.1148. EMBO J. 2002. PMID: 11867543 Free PMC article.
-
Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge.Trends Biochem Sci. 1997 Apr;22(4):132-7. doi: 10.1016/s0968-0004(97)01018-9. Trends Biochem Sci. 1997. PMID: 9149533 Review.
-
CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast.Curr Opin Struct Biol. 2016 Feb;36:48-57. doi: 10.1016/j.sbi.2015.12.005. Epub 2016 Jan 21. Curr Opin Struct Biol. 2016. PMID: 26803803 Free PMC article. Review.
Cited by
-
A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing.PLoS Genet. 2019 Oct 31;15(10):e1008460. doi: 10.1371/journal.pgen.1008460. eCollection 2019 Oct. PLoS Genet. 2019. PMID: 31671093 Free PMC article.
-
Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions.Mol Cell Biol. 2002 May;22(10):3219-29. doi: 10.1128/MCB.22.10.3219-3229.2002. Mol Cell Biol. 2002. PMID: 11971955 Free PMC article.
-
Transcriptome analysis of the zebrafish mind bomb mutant.Mol Genet Genomics. 2009 Jan;281(1):77-85. doi: 10.1007/s00438-008-0395-5. Epub 2008 Nov 13. Mol Genet Genomics. 2009. PMID: 19005681
-
Stable tri-snRNP integration is accompanied by a major structural rearrangement of the spliceosome that is dependent on Prp8 interaction with the 5' splice site.RNA. 2015 Nov;21(11):1993-2005. doi: 10.1261/rna.053991.115. Epub 2015 Sep 18. RNA. 2015. PMID: 26385511 Free PMC article.
-
Fragile X mental retardation protein recognizes a G quadruplex structure within the survival motor neuron domain containing 1 mRNA 5'-UTR.Mol Biosyst. 2017 Jul 25;13(8):1448-1457. doi: 10.1039/c7mb00070g. Mol Biosyst. 2017. PMID: 28612854 Free PMC article.
References
-
- Behrens S.E. and Lührmann,R. (1991) Immunoaffinity purification of a U4/U6⋅U5 tri-snRNP from human cells. Genes Dev., 5, 1439–1452. - PubMed
-
- Behrens S.E., Gallison,F., Legrain,P. and Lührmann,R. (1993a) Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosome. Proc. Natl Acad. Sci. USA, 90, 8229–8233. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

