Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs
- PMID: 11711622
- PMCID: PMC116128
- DOI: 10.1128/JVI.75.24.12319-12330.2001
Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs
Abstract
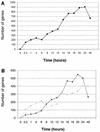
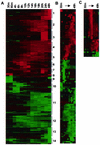
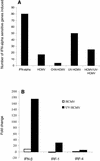
The effect of human cytomegalovirus (HCMV) infection on cellular mRNA accumulation was analyzed by gene chip technology. During a 48-h time course after infection of human diploid fibroblasts, 1,425 cellular mRNAs were found to be up-regulated or down-regulated by threefold or greater in at least two consecutive time points. Several classes of genes were prominently affected, including interferon response genes, cell cycle regulators, apoptosis regulators, inflammatory pathway genes, and immune regulators. The number of mRNAs that were up-regulated or down-regulated were roughly equal over the complete time course. However, for the first 8 h after infection, the number of up-regulated mRNAs was significantly less than the number of down-regulated mRNAs. By analyzing the mRNA expression profile of cells infected in the presence of cycloheximide, it was found that a minimum of 25 mRNAs were modulated by HCMV in the absence of protein synthesis. These included mRNAs encoded by a small number of interferon-responsive genes, as well as beta interferon itself. Cellular mRNA levels in cytomegalovirus-infected cells were compared to the levels in cells infected with UV-inactivated virus. The inactivated virus caused the up-regulation of a much greater number of mRNAs, many of which encoded proteins with antiviral roles, such as interferon-responsive genes and proinflammatory cytokines. These data argue that one or more newly synthesized viral gene products block the induction of antiviral pathways that are triggered by HCMV binding and entry.
Figures

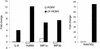
Similar articles
-
Processing bodies accumulate in human cytomegalovirus-infected cells and do not affect viral replication at high multiplicity of infection.Virology. 2014 Jun;458-459:151-61. doi: 10.1016/j.virol.2014.04.022. Epub 2014 May 13. Virology. 2014. PMID: 24928047
-
Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs.Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13985-90. doi: 10.1073/pnas.94.25.13985. Proc Natl Acad Sci U S A. 1997. PMID: 9391139 Free PMC article.
-
Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA.Gene. 2014 Feb 25;536(2):272-8. doi: 10.1016/j.gene.2013.12.012. Epub 2013 Dec 18. Gene. 2014. PMID: 24361963
-
Posttranscriptional suppression of interleukin-6 production by human cytomegalovirus.J Virol. 2005 Jan;79(1):472-85. doi: 10.1128/JVI.79.1.472-485.2005. J Virol. 2005. PMID: 15596840 Free PMC article.
-
[Interrelationship between human cytomegalovirus infection and chemokine].Nihon Rinsho. 1998 Jan;56(1):69-74. Nihon Rinsho. 1998. PMID: 9465667 Review. Japanese.
Cited by
-
The conserved Cockayne syndrome B-piggyBac fusion protein (CSB-PGBD3) affects DNA repair and induces both interferon-like and innate antiviral responses in CSB-null cells.DNA Repair (Amst). 2012 May 1;11(5):488-501. doi: 10.1016/j.dnarep.2012.02.004. Epub 2012 Apr 6. DNA Repair (Amst). 2012. PMID: 22483866 Free PMC article.
-
Human cytomegalovirus inhibits apoptosis by proteasome-mediated degradation of Bax at endoplasmic reticulum-mitochondrion contacts.J Virol. 2013 May;87(10):5657-68. doi: 10.1128/JVI.00145-13. Epub 2013 Mar 13. J Virol. 2013. PMID: 23487455 Free PMC article.
-
Increased expression of LDL receptor-related protein 1 during human cytomegalovirus infection reduces virion cholesterol and infectivity.Cell Host Microbe. 2012 Jul 19;12(1):86-96. doi: 10.1016/j.chom.2012.05.012. Cell Host Microbe. 2012. PMID: 22817990 Free PMC article.
-
Human cytomegalovirus harnesses host L1 retrotransposon for efficient replication.Nat Commun. 2024 Sep 2;15(1):7640. doi: 10.1038/s41467-024-51961-y. Nat Commun. 2024. PMID: 39223139 Free PMC article.
-
Transcriptome signature of virulent and attenuated pseudorabies virus-infected rodent brain.J Virol. 2006 Feb;80(4):1773-86. doi: 10.1128/JVI.80.4.1773-1786.2006. J Virol. 2006. PMID: 16439534 Free PMC article.
References
-
- Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P, Dolinski K, Dwight S S, Eppig J T, Harris M A, Hill D P, Issel-Tarver L, Kasarskis A, Lewis S, Matese J C, Richardson J E, Ringwald M, Rubin G M, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
-
- Bozza M, Bliss J L, Dorner A J, Trepicchio W L. Interleukin-11 modulates Th1/Th2 cytokine production from activated CD4+ T cells. J Interferon Cytokine Res. 2001;21:21–30. - PubMed
-
- Bresnahan W A, Bolldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

