Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome
- PMID: 11805330
- PMCID: PMC117378
- DOI: 10.1073/pnas.231608898
Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome
Abstract
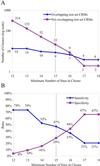
A major challenge in interpreting genome sequences is understanding how the genome encodes the information that specifies when and where a gene will be expressed. The first step in this process is the identification of regions of the genome that contain regulatory information. In higher eukaryotes, this cis-regulatory information is organized into modular units [cis-regulatory modules (CRMs)] of a few hundred base pairs. A common feature of these cis-regulatory modules is the presence of multiple binding sites for multiple transcription factors. Here, we evaluate the extent to which the tendency for transcription factor binding sites to be clustered can be used as the basis for the computational identification of cis-regulatory modules. By using published DNA binding specificity data for five transcription factors active in the early Drosophila embryo, we identified genomic regions containing unusually high concentrations of predicted binding sites for these factors. A significant fraction of these binding site clusters overlap known CRMs that are regulated by these factors. In addition, many of the remaining clusters are adjacent to genes expressed in a pattern characteristic of genes regulated by these factors. We tested one of the newly identified clusters, mapping upstream of the gap gene giant (gt), and show that it acts as an enhancer that recapitulates the posterior expression pattern of gt.
Figures


Comment in
-
Deciphering genetic regulatory codes: a challenge for functional genomics.Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):546-8. doi: 10.1073/pnas.032685999. Proc Natl Acad Sci U S A. 2002. PMID: 11805309 Free PMC article. No abstract available.
Similar articles
-
Decoding cis-regulatory DNAs in the Drosophila genome.Curr Opin Genet Dev. 2002 Oct;12(5):601-6. doi: 10.1016/s0959-437x(02)00345-3. Curr Opin Genet Dev. 2002. PMID: 12200166 Review.
-
Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo.BMC Bioinformatics. 2002 Oct 24;3:30. doi: 10.1186/1471-2105-3-30. Epub 2002 Oct 24. BMC Bioinformatics. 2002. PMID: 12398796 Free PMC article.
-
Transcriptional control in the segmentation gene network of Drosophila.PLoS Biol. 2004 Sep;2(9):E271. doi: 10.1371/journal.pbio.0020271. Epub 2004 Aug 31. PLoS Biol. 2004. PMID: 15340490 Free PMC article.
-
De novo prediction of cis-regulatory elements and modules through integrative analysis of a large number of ChIP datasets.BMC Genomics. 2014 Dec 2;15:1047. doi: 10.1186/1471-2164-15-1047. BMC Genomics. 2014. PMID: 25442502 Free PMC article.
-
Organizing combinatorial transcription factor recruitment at cis-regulatory modules.Transcription. 2018;9(4):233-239. doi: 10.1080/21541264.2017.1394424. Epub 2017 Nov 28. Transcription. 2018. PMID: 29105538 Free PMC article. Review.
Cited by
-
Tandem machine learning for the identification of genes regulated by transcription factors.BMC Bioinformatics. 2005 Aug 22;6:204. doi: 10.1186/1471-2105-6-204. BMC Bioinformatics. 2005. PMID: 16115317 Free PMC article.
-
CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling.Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12114-9. doi: 10.1073/pnas.0402858101. Epub 2004 Aug 5. Proc Natl Acad Sci U S A. 2004. PMID: 15297614 Free PMC article.
-
Some statistical properties of regulatory DNA sequences, and their use in predicting regulatory regions in the Drosophila genome: the fluffy-tail test.BMC Bioinformatics. 2005 Apr 27;6:109. doi: 10.1186/1471-2105-6-109. BMC Bioinformatics. 2005. PMID: 15857505 Free PMC article.
-
Reliable prediction of regulator _targets using 12 Drosophila genomes.Genome Res. 2007 Dec;17(12):1919-31. doi: 10.1101/gr.7090407. Epub 2007 Nov 7. Genome Res. 2007. PMID: 17989251 Free PMC article.
-
An in silico analysis of robust but fragile gene regulation links enhancer length to robustness.PLoS Comput Biol. 2019 Nov 15;15(11):e1007497. doi: 10.1371/journal.pcbi.1007497. eCollection 2019 Nov. PLoS Comput Biol. 2019. PMID: 31730659 Free PMC article.
References
-
- Carroll S B, Grenier J K, Weatherbee S D. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Oxford: Blackwell Scientific; 2001.
-
- Goto T, Macdonald P, Maniatis T. Cell. 1989;57:413–422. - PubMed
-
- Stanojevic D, Small S, Levine M. Science. 1991;254:1385–1387. - PubMed
-
- Small S, Blair A, Levine M. Dev Biol. 1996;175:314–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

