Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation
- PMID: 11880622
- PMCID: PMC122407
- DOI: 10.1073/pnas.012027399
Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation
Abstract
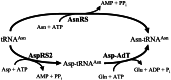
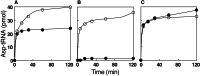
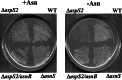
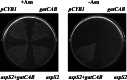
Biochemical experiments and genomic sequence analysis showed that Deinococcus radiodurans and Thermus thermophilus do not possess asparagine synthetase (encoded by asnA or asnB), the enzyme forming asparagine from aspartate. Instead these organisms derive asparagine from asparaginyl-tRNA, which is made from aspartate in the tRNA-dependent transamidation pathway [Becker, H. D. & Kern, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12832-12837; and Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Söll, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12838-12843]. A genetic knockout disrupting this pathway deprives D. radiodurans of the ability to synthesize asparagine and confers asparagine auxotrophy. The organism's capacity to make asparagine could be restored by transformation with Escherichia coli asnB. This result demonstrates that in Deinococcus, the only route to asparagine is via asparaginyl-tRNA. Analysis of the completed genomes of many bacteria reveal that, barring the existence of an unknown pathway of asparagine biosynthesis, a wide spectrum of bacteria rely on the tRNA-dependent transamidation pathway as the sole route to asparagine.
Figures
Similar articles
-
From one amino acid to another: tRNA-dependent amino acid biosynthesis.Nucleic Acids Res. 2008 Apr;36(6):1813-25. doi: 10.1093/nar/gkn015. Epub 2008 Feb 5. Nucleic Acids Res. 2008. PMID: 18252769 Free PMC article. Review.
-
Isolation, crystallization and preliminary X-ray analysis of the transamidosome, a ribonucleoprotein involved in asparagine formation.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Jun 1;65(Pt 6):577-81. doi: 10.1107/S1744309109015000. Epub 2009 May 22. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19478435 Free PMC article.
-
The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid.EMBO J. 1998 May 15;17(10):2947-60. doi: 10.1093/emboj/17.10.2947. EMBO J. 1998. PMID: 9582288 Free PMC article.
-
Glutamyl-tRNA(Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis.Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):12838-43. doi: 10.1073/pnas.95.22.12838. Proc Natl Acad Sci U S A. 1998. PMID: 9789001 Free PMC article.
-
Aminoacyl-tRNA synthesis by pre-translational amino acid modification.RNA Biol. 2004 May;1(1):16-20. Epub 2004 May 28. RNA Biol. 2004. PMID: 17194933 Review.
Cited by
-
The genome of Pelobacter carbinolicus reveals surprising metabolic capabilities and physiological features.BMC Genomics. 2012 Dec 10;13:690. doi: 10.1186/1471-2164-13-690. BMC Genomics. 2012. PMID: 23227809 Free PMC article.
-
Emergence and evolution.Top Curr Chem. 2014;344:43-87. doi: 10.1007/128_2013_423. Top Curr Chem. 2014. PMID: 23478877 Free PMC article. Review.
-
The archaeal transamidosome for RNA-dependent glutamine biosynthesis.Nucleic Acids Res. 2010 Sep;38(17):5774-83. doi: 10.1093/nar/gkq336. Epub 2010 May 10. Nucleic Acids Res. 2010. PMID: 20457752 Free PMC article.
-
Assays for transfer RNA-dependent amino acid biosynthesis.Methods. 2008 Feb;44(2):139-45. doi: 10.1016/j.ymeth.2007.06.010. Methods. 2008. PMID: 18241795 Free PMC article.
-
From one amino acid to another: tRNA-dependent amino acid biosynthesis.Nucleic Acids Res. 2008 Apr;36(6):1813-25. doi: 10.1093/nar/gkn015. Epub 2008 Feb 5. Nucleic Acids Res. 2008. PMID: 18252769 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

