A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry
- PMID: 12237468
- PMCID: PMC2373692
- DOI: 10.1110/ps.0212602
A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry
Abstract
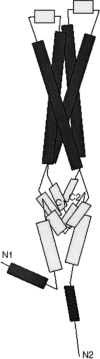
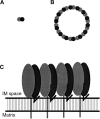
The mitochondrial prohibitin complex consists of two subunits (PHB1 of 32 kD and PHB2 of 34 kD), assembled into a membrane-associated supercomplex of approximately 1 MD. A chaperone-like function in holding and assembling newly synthesized mitochondrial polypeptide chains has been proposed. To further elucidate the function of this complex, structural information is necessary. In this study we use chemical crosslinking, connecting lysine side chains, which are well scattered along the sequence. Crosslinked peptides from protease digested prohibitin complexes were identified with mass spectrometry. From these results, spatial restraints for possible protein conformation were obtained. Many interaction sites between PHB1 and PHB2 were found, whereas no homodimeric interactions were observed. Secondary and tertiary structural predictions were made using several algorithms and the models best fitting the spatial restraints were selected for further evaluation. From the structure predictions and the crosslink data we derived a structural building block of one PHB1 and one PHB2 subunit, strongly intertwined along most of their length. The size of the complex implies that approximately 14 of these building blocks are present. Each unit contains a putative transmembrane helix in PHB2. Taken together with the unit building block we postulate a circular palisade-like arrangement of the building blocks projecting into the intermembrane space.
Figures

Similar articles
-
Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins.EMBO J. 2000 Jun 1;19(11):2444-51. doi: 10.1093/emboj/19.11.2444. EMBO J. 2000. PMID: 10835343 Free PMC article.
-
Formation of membrane-bound ring complexes by prohibitins in mitochondria.Mol Biol Cell. 2005 Jan;16(1):248-59. doi: 10.1091/mbc.e04-09-0807. Epub 2004 Nov 3. Mol Biol Cell. 2005. PMID: 15525670 Free PMC article.
-
Prohibitin and mitochondrial biology.Trends Endocrinol Metab. 2009 Oct;20(8):394-401. doi: 10.1016/j.tem.2009.04.004. Epub 2009 Sep 3. Trends Endocrinol Metab. 2009. PMID: 19733482 Review.
-
Functional studies of Plasmodium falciparum's prohibitin1 and prohibitin 2 in yeast.Indian J Med Microbiol. 2020 Apr-Jun;38(2):213-215. doi: 10.4103/ijmm.IJMM_20_28. Indian J Med Microbiol. 2020. PMID: 32883936
-
Prohibitin 2: At a communications crossroads.IUBMB Life. 2015 Apr;67(4):239-54. doi: 10.1002/iub.1366. Epub 2015 Apr 21. IUBMB Life. 2015. PMID: 25904163 Review.
Cited by
-
Analysis of the effect of the mitochondrial prohibitin complex, a context-dependent modulator of longevity, on the C. elegans metabolome.Biochim Biophys Acta. 2015 Nov;1847(11):1457-68. doi: 10.1016/j.bbabio.2015.06.003. Epub 2015 Jun 17. Biochim Biophys Acta. 2015. PMID: 26092086 Free PMC article.
-
Dysfunction of Prohibitin 2 Results in Reduced Susceptibility to Multiple Antifungal Drugs via Activation of the Oxidative Stress-Responsive Transcription Factor Pap1 in Fission Yeast.Antimicrob Agents Chemother. 2018 Oct 24;62(11):e00860-18. doi: 10.1128/AAC.00860-18. Print 2018 Nov. Antimicrob Agents Chemother. 2018. PMID: 30181366 Free PMC article.
-
Response to "Comment on 'A method to measure cellular adhesion utilizing a polymer micro-cantilever'" [Appl. Phys. Lett. 104, 236103 (2014)].Appl Phys Lett. 2014 Jun 9;104(23):236104. doi: 10.1063/1.4882185. Epub 2014 Jun 10. Appl Phys Lett. 2014. PMID: 25315106 Free PMC article. No abstract available.
-
Inhibition of insulin/IGF-1 receptor signaling protects from mitochondria-mediated kidney failure.EMBO Mol Med. 2015 Mar;7(3):275-87. doi: 10.15252/emmm.201404916. EMBO Mol Med. 2015. PMID: 25643582 Free PMC article.
-
Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1.J Biol Chem. 2010 Apr 9;285(15):11445-57. doi: 10.1074/jbc.M109.065425. Epub 2010 Feb 11. J Biol Chem. 2010. PMID: 20150421 Free PMC article.
References
-
- Back, J.W., Hartog, A.F., Dekker, H.L., Muijsers, A.O., de Koning. L.J., and de Jong, L. 2001. A new cross-linker for mass spectrometric analysis of the quaternary structure of protein complexes. J. Am. Soc. Mass Spectrom. 12 222–227. - PubMed
-
- Bennett, K.L., Kussmann, M., Bjork, P., Godzwon, M., Mikkelsen, M., Sorensen, P., and Roepstorff, P. 2000a. Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping—A novel approach to assess intermolecular protein contacts. Protein Sci. 9 1503–1518. - PMC - PubMed
-
- Bennett, K.L., Matthiesen, T., and Roepstorff, P. 2000b. Probing protein surface topology by chemical surface labeling, cross-linking, and mass spectrometry. Methods Mol. Biol. 146 113–131. - PubMed
-
- Coates, P.J., Jamieson, D.J., Smart, K., Prescott, A.R., and Hall, P.A. 1997. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr. Biol. 7 607–610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

