Structural analysis of the RSC chromatin-remodeling complex
- PMID: 12368485
- PMCID: PMC129698
- DOI: 10.1073/pnas.162504299
Structural analysis of the RSC chromatin-remodeling complex
Abstract
Electron microscopy of the RSC chromatin-remodeling complex reveals a ring of protein densities around a central cavity. The size and shape of the cavity correspond closely to those of a nucleosome. Results of nuclease protection analysis are consistent with nucleosome binding in the cavity. Such binding could explain the ability of RSC to expose nucleosomal DNA in the presence of ATP without loss of associated histones.
Figures

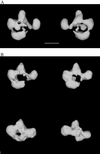
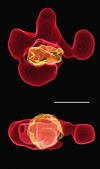

Similar articles
-
Electron microscopic analysis of the RSC chromatin remodeling complex.Methods Enzymol. 2004;376:48-62. doi: 10.1016/S0076-6879(03)76004-2. Methods Enzymol. 2004. PMID: 14975298 Review. No abstract available.
-
Structure of a RSC-nucleosome complex and insights into chromatin remodeling.Nat Struct Mol Biol. 2008 Dec;15(12):1272-7. doi: 10.1038/nsmb.1524. Epub 2008 Nov 23. Nat Struct Mol Biol. 2008. PMID: 19029894 Free PMC article.
-
Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.Elife. 2019 Dec 30;8:e54449. doi: 10.7554/eLife.54449. Elife. 2019. PMID: 31886770 Free PMC article.
-
Histone octamer transfer by a chromatin-remodeling complex.Cell. 1999 Feb 5;96(3):389-92. doi: 10.1016/s0092-8674(00)80551-6. Cell. 1999. PMID: 10025404
-
ATP-dependent chromatin remodeling and DNA double-strand break repair.Cell Cycle. 2005 Aug;4(8):1011-4. doi: 10.4161/cc.4.8.1887. Epub 2005 Aug 2. Cell Cycle. 2005. PMID: 16082209 Review.
Cited by
-
A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding.Cell. 2010 Apr 30;141(3):407-18. doi: 10.1016/j.cell.2010.03.048. Cell. 2010. PMID: 20434983 Free PMC article.
-
MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF.Biochemistry. 2008 Dec 23;47(51):13726-32. doi: 10.1021/bi8016944. Biochemistry. 2008. PMID: 19035833 Free PMC article.
-
Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events.J Biol Chem. 2011 Dec 2;286(48):41883-41892. doi: 10.1074/jbc.M111.301465. Epub 2011 Oct 3. J Biol Chem. 2011. PMID: 21969370 Free PMC article.
-
Architecture of the SWI/SNF-nucleosome complex.Mol Cell Biol. 2008 Oct;28(19):6010-21. doi: 10.1128/MCB.00693-08. Epub 2008 Jul 21. Mol Cell Biol. 2008. PMID: 18644858 Free PMC article.
-
Mechanisms for ATP-dependent chromatin remodelling: the means to the end.FEBS J. 2011 Oct;278(19):3579-95. doi: 10.1111/j.1742-4658.2011.08281.x. Epub 2011 Sep 8. FEBS J. 2011. PMID: 21810178 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

