A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition
- PMID: 12461076
- PMCID: PMC2194258
- DOI: 10.1084/jem.20020797
A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition
Abstract
Human histocompatibility leukocyte antigen (HLA)-E is a nonclassical major histocompatibility complex (MHC) class I molecule which presents a restricted set of nonameric peptides, derived mainly from the signal sequence of other MHC class I molecules. It interacts with CD94/NKG2 receptors expressed on the surface of natural killer (NK) cells and T cell subsets. Here we demonstrate that HLA-E also presents a peptide derived from the leader sequence of human heat shock protein 60 (hsp60). This peptide gains access to HLA-E intracellularly, resulting in up-regulated HLA-E/hsp60 signal peptide cell-surface levels on stressed cells. Notably, HLA-E molecules in complex with the hsp60 signal peptide are no longer recognized by CD94/NKG2A inhibitory receptors. Thus, during cellular stress an increased proportion of HLA-E molecules may bind the nonprotective hsp60 signal peptide, leading to a reduced capacity to inhibit a major NK cell population. Such stress induced peptide interference would gradually uncouple CD94/NKG2A inhibitory recognition and provide a mechanism for NK cells to detect stressed cells in a peptide-dependent manner.
Figures

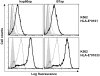
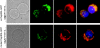

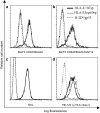
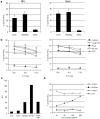
Comment in
-
Stress signals activate natural killer cells.J Exp Med. 2002 Dec 2;196(11):1399-402. doi: 10.1084/jem.20021747. J Exp Med. 2002. PMID: 12461075 Free PMC article. No abstract available.
Similar articles
-
HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C.Nature. 1998 Feb 19;391(6669):795-9. doi: 10.1038/35869. Nature. 1998. PMID: 9486650
-
Specific recognition of HLA-E, but not classical, HLA class I molecules by soluble CD94/NKG2A and NK cells.J Immunol. 1999 Jan 1;162(1):305-13. J Immunol. 1999. PMID: 9886400
-
Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E.EMBO J. 1999 Aug 2;18(15):4250-60. doi: 10.1093/emboj/18.15.4250. EMBO J. 1999. PMID: 10428963 Free PMC article.
-
Natural killer cell surveillance of intracellular antigen processing pathways mediated by recognition of HLA-E and Qa-1b by CD94/NKG2 receptors.Microbes Infect. 2000 Apr;2(4):371-80. doi: 10.1016/s1286-4579(00)00330-0. Microbes Infect. 2000. PMID: 10817639 Review.
-
Natural killer cell recognition of HLA class I molecules.Rev Immunogenet. 2000;2(3):433-48. Rev Immunogenet. 2000. PMID: 11256749 Review.
Cited by
-
Deciphering the HLA-E immunopeptidome with mass spectrometry: an opportunity for universal mRNA vaccines and T-cell-directed immunotherapies.Front Immunol. 2024 Sep 5;15:1442783. doi: 10.3389/fimmu.2024.1442783. eCollection 2024. Front Immunol. 2024. PMID: 39301027 Free PMC article. Review.
-
Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes.Genome Res. 2009 May;19(5):757-69. doi: 10.1101/gr.085738.108. Genome Res. 2009. PMID: 19411600 Free PMC article.
-
Opportunities and limitations of natural killer cells as adoptive therapy for malignant disease.Cytotherapy. 2014 Nov;16(11):1453-1466. doi: 10.1016/j.jcyt.2014.03.009. Epub 2014 May 20. Cytotherapy. 2014. PMID: 24856895 Free PMC article. Review.
-
Activating KIRs and NKG2C in Viral Infections: Toward NK Cell Memory?Front Immunol. 2015 Nov 9;6:573. doi: 10.3389/fimmu.2015.00573. eCollection 2015. Front Immunol. 2015. PMID: 26617607 Free PMC article. Review.
-
Non-Classical HLA Class 1b and Hepatocellular Carcinoma.Biomedicines. 2023 Jun 9;11(6):1672. doi: 10.3390/biomedicines11061672. Biomedicines. 2023. PMID: 37371767 Free PMC article. Review.
References
-
- Lanier, L.L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:359–393. - PubMed
-
- Drickamer, K. 1999. C-type lectin-like domains. Curr. Opin. Struct. Biol. 9:585–590. - PubMed
-
- Boyington, J.C., A.G. Brooks, and P.D. Sun. 2001. Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol. Rev. 181:66–78. - PubMed
-
- Braud, V.M., D.S. Allan, C.A. O'Callaghan, K. Söderström, A. D'Andrea, G.S. Ogg, S. Lazetic, N.T. Young, J.I. Bell, J.H. Phillips, et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 391:795–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

