Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis
- PMID: 12651943
- PMCID: PMC153039
- DOI: 10.1073/pnas.0730870100
Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis
Abstract
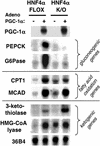
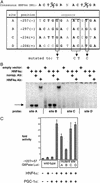
The liver plays several critical roles in the metabolic adaptation to fasting. We have shown previously that the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha) is induced in fasted or diabetic liver and activates the entire program of gluconeogenesis. PGC-1alpha interacts with several nuclear receptors known to bind gluconeogenic promoters including the glucocorticoid receptor, hepatocyte nuclear factor 4alpha (HNF4alpha), and the peroxisome proliferator-activated receptors. However, the genetic requirement for any of these interactions has not been determined. Using hepatocytes from mice lacking HNF4alpha in the liver, we show here that PGC-1alpha completely loses its ability to activate key genes of gluconeogenesis such as phosphoenolpyruvate carboxykinase and glucose-6-phosphatase when HNF4alpha is absent. It is also shown that PGC-1alpha can induce genes of beta-oxidation and ketogenesis in hepatocytes, but these effects do not require HNF4alpha. Analysis of the glucose-6-phosphatase promoter indicates a key role for HNF4alpha-binding sites that function robustly only when HNF4alpha is coactivated by PGC-1alpha. These data illustrate the involvement of PGC-1alpha in several aspects of the hepatic fasting response and show that HNF4alpha is a critical component of PGC-1alpha-mediated gluconeogenesis.
Figures


Similar articles
-
Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1.Nature. 2001 Sep 13;413(6852):131-8. doi: 10.1038/35093050. Nature. 2001. PMID: 11557972
-
Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR.Genes Dev. 2004 Jan 15;18(2):157-69. doi: 10.1101/gad.1138104. Epub 2004 Jan 16. Genes Dev. 2004. PMID: 14729567 Free PMC article.
-
Peroxisome proliferator-activated receptor gamma coactivator-1alpha, as a transcription amplifier, is not essential for basal and hormone-induced phosphoenolpyruvate carboxykinase gene expression.Mol Endocrinol. 2004 Apr;18(4):807-19. doi: 10.1210/me.2003-0384. Mol Endocrinol. 2004. PMID: 15044597
-
Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis.Int J Biochem Cell Biol. 2012 Jan;44(1):33-45. doi: 10.1016/j.biocel.2011.10.001. Epub 2011 Oct 8. Int J Biochem Cell Biol. 2012. PMID: 22004992 Review.
-
Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha.Int J Obes (Lond). 2005 Mar;29 Suppl 1:S5-9. doi: 10.1038/sj.ijo.0802905. Int J Obes (Lond). 2005. PMID: 15711583 Review.
Cited by
-
Liver-Specific Knockdown of Class IIa HDACs Has Limited Efficacy on Glucose Metabolism but Entails Severe Organ Side Effects in Mice.Front Endocrinol (Lausanne). 2020 Aug 28;11:598. doi: 10.3389/fendo.2020.00598. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 32982982 Free PMC article.
-
MAPK phosphatase-3 promotes hepatic gluconeogenesis through dephosphorylation of forkhead box O1 in mice.J Clin Invest. 2010 Nov;120(11):3901-11. doi: 10.1172/JCI43250. J Clin Invest. 2010. PMID: 20921625 Free PMC article.
-
Exploring the signal-dependent transcriptional regulation involved in the liver pathology of type 2 diabetes.Diabetol Int. 2022 Dec 3;14(1):15-20. doi: 10.1007/s13340-022-00610-0. eCollection 2023 Jan. Diabetol Int. 2022. PMID: 36636166 Free PMC article. Review.
-
Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue.Diabetes. 2010 Jun;59(6):1338-48. doi: 10.2337/db09-1324. Epub 2010 Mar 18. Diabetes. 2010. PMID: 20299475 Free PMC article.
-
Regulation of carnitine palmitoyltransferase I (CPT-Ialpha) gene expression by the peroxisome proliferator activated receptor gamma coactivator (PGC-1) isoforms.Mol Cell Endocrinol. 2007 Mar 15;267(1-2):6-16. doi: 10.1016/j.mce.2006.11.012. Epub 2006 Dec 16. Mol Cell Endocrinol. 2007. PMID: 17239528 Free PMC article.
References
-
- Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. Cell. 1998;92:829–839. - PubMed
-
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Cell. 1999;98:115–124. - PubMed
-
- Lin J, Wu H, Tarr P T, Zhang C Y, Wu Z, Boss O, Michael L F, Puigserver P, Isotani E, Olson E N, et al. Nature. 2002;418:797–801. - PubMed
-
- Yoon J C, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn C R, Granner D K, et al. Nature. 2001;413:131–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

