The G protein-coupled receptor repertoires of human and mouse
- PMID: 12679517
- PMCID: PMC153653
- DOI: 10.1073/pnas.0230374100
The G protein-coupled receptor repertoires of human and mouse
Abstract
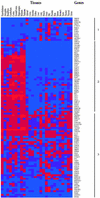
Diverse members of the G protein-coupled receptor (GPCR) superfamily participate in a variety of physiological functions and are major _targets of pharmaceutical drugs. Here we report that the repertoire of GPCRs for endogenous ligands consists of 367 receptors in humans and 392 in mice. Included here are 26 human and 83 mouse GPCRs not previously identified. A direct comparison of GPCRs in the two species reveals an unexpected level of orthology. The evolutionary preservation of these molecules argues against functional redundancy among highly related receptors. Phylogenetic analyses cluster 60% of GPCRs according to ligand preference, allowing prediction of ligand types for dozens of orphan receptors. Expression profiling of 100 GPCRs demonstrates that most are expressed in multiple tissues and that individual tissues express multiple GPCRs. Over 90% of GPCRs are expressed in the brain. Strikingly, however, the profiles of most GPCRs are unique, yielding thousands of tissue- and cell-specific receptor combinations for the modulation of physiological processes.
Figures



Similar articles
-
Relaxin-3, INSL5, and their receptors.Results Probl Cell Differ. 2008;46:213-37. doi: 10.1007/400_2007_055. Results Probl Cell Differ. 2008. PMID: 18236022 Review.
-
Molecular cloning and characterization of two novel retinoic acid-inducible orphan G-protein-coupled receptors (GPRC5B and GPRC5C).Genomics. 2000 Jul 1;67(1):8-18. doi: 10.1006/geno.2000.6226. Genomics. 2000. PMID: 10945465
-
The G protein-coupled receptor subset of the rat genome.BMC Genomics. 2007 Sep 25;8:338. doi: 10.1186/1471-2164-8-338. BMC Genomics. 2007. PMID: 17892602 Free PMC article.
-
Proenkephalin A gene products activate a new family of sensory neuron--specific GPCRs.Nat Neurosci. 2002 Mar;5(3):201-9. doi: 10.1038/nn815. Nat Neurosci. 2002. PMID: 11850634
-
Orphan G-protein-coupled receptors: the next generation of drug _targets?Br J Pharmacol. 1998 Dec;125(7):1387-92. doi: 10.1038/sj.bjp.0702238. Br J Pharmacol. 1998. PMID: 9884064 Free PMC article. Review.
Cited by
-
Novel computational methodologies for structural modeling of spacious ligand binding sites of G-protein-coupled receptors: development and application to human leukotriene B4 receptor.ScientificWorldJournal. 2012;2012:691579. doi: 10.1100/2012/691579. Epub 2012 Dec 10. ScientificWorldJournal. 2012. PMID: 23304088 Free PMC article.
-
A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2.Antiviral Res. 2012 Jun;94(3):288-96. doi: 10.1016/j.antiviral.2011.12.012. Epub 2012 Jan 17. Antiviral Res. 2012. PMID: 22265858 Free PMC article.
-
The chemokine superfamily revisited.Immunity. 2012 May 25;36(5):705-16. doi: 10.1016/j.immuni.2012.05.008. Immunity. 2012. PMID: 22633458 Free PMC article. Review.
-
Procedure for calculation of potency and efficacy for ligands acting on Gs- and Gi-coupled receptors.Neurochem Res. 2012 Dec;37(12):2767-75. doi: 10.1007/s11064-012-0870-6. Epub 2012 Sep 26. Neurochem Res. 2012. PMID: 23011203
-
Unsung renal receptors: orphan G-protein-coupled receptors play essential roles in renal development and homeostasis.Acta Physiol (Oxf). 2017 Jun;220(2):189-200. doi: 10.1111/apha.12813. Epub 2016 Oct 23. Acta Physiol (Oxf). 2017. PMID: 27699982 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

