Tandem tetramer-based microsatellite fingerprinting for typing of Proteus mirabilis strains
- PMID: 12682159
- PMCID: PMC153880
- DOI: 10.1128/JCM.41.4.1673-1680.2003
Tandem tetramer-based microsatellite fingerprinting for typing of Proteus mirabilis strains
Abstract
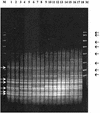
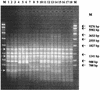
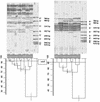
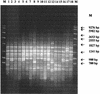
Two microsatellite tandem repeated tetramers, (GACA)(4) and (CAAT)(4), were used for Proteus mirabilis strain differentiation. The microsatellite-based PCR tests were applied for the examination of interstrain diversity for 87 P. mirabilis strains. Forty-six of the investigated strains were clinical isolates (5 were hospital isolates and 39 were outpatient clinic isolates); 42 strains were derived from the Kauffmann-Perch collection of laboratory strains. Fingerprinting done with the tetramers had a high discrimination ability [0.992 and 0.940 for (GACA)(4) and (CAAT)(4), respectively]. The distributions of clinical isolates among well-defined laboratory strains, determined by numerical analysis (unweighted pair-group method with arithmetic averages; Dice similarity coefficient), proved their genetic similarity to reference strains in the Kauffmann-Perch collection. This analysis also indicated that it is possible to estimate some phenotypic properties of P. mirabilis clinical isolates solely on the basis of microsatellite fingerprinting.
Figures

Similar articles
-
Arbitrarily primed polymerase chain reaction provides rapid differentiation of Proteus mirabilis isolates from a pediatric hospital.J Clin Microbiol. 1993 May;31(5):1055-9. doi: 10.1128/jcm.31.5.1055-1059.1993. J Clin Microbiol. 1993. PMID: 8099079 Free PMC article.
-
Comparison of PCR-based molecular markers for the characterization of Proteus mirabilis clinical isolates.Braz J Infect Dis. 2008 Oct;12(5):423-9. doi: 10.1590/s1413-86702008000500014. Braz J Infect Dis. 2008. PMID: 19219283
-
Spread of multidrug-resistant Proteus mirabilis isolates producing an AmpC-type beta-lactamase: epidemiology and clinical management.Int J Antimicrob Agents. 2009 Apr;33(4):328-33. doi: 10.1016/j.ijantimicag.2008.09.007. Epub 2008 Dec 17. Int J Antimicrob Agents. 2009. PMID: 19095415
-
A novel functional class 2 integron in clinical Proteus mirabilis isolates.J Antimicrob Chemother. 2014 Apr;69(4):973-6. doi: 10.1093/jac/dkt456. Epub 2013 Nov 13. J Antimicrob Chemother. 2014. PMID: 24235093
-
Molecular fingerprints to identify Candida species.Biomed Res Int. 2013;2013:923742. doi: 10.1155/2013/923742. Epub 2013 Jun 17. Biomed Res Int. 2013. PMID: 23844370 Free PMC article. Review.
Cited by
-
Developing lactic acid bacteria starter cultures for wholemeal rye flour bread with improved functionality, nutritional value, taste, appearance and safety.PLoS One. 2022 Jan 14;17(1):e0261677. doi: 10.1371/journal.pone.0261677. eCollection 2022. PLoS One. 2022. PMID: 35030182 Free PMC article.
References
-
- Arribas, R., S. Tortola, J. Welsh, M. McClelland, and M. Peinado. 1996. Arbitrarily primed PCR and RAPDs, p. 47-53. In M. R. Micheli and R. Bova (ed.), Fingerprinting methods based on arbitrarily primed PCR. Springer-Verlag KG, Berlin, Germany.
-
- Cieślikowski, T., D. Gradecka, M. Mielczarek, and W. Kaca. 2000. Different PCR-based fingerprints exhibit significantly higher intra-serotype genetic similarity for P. mirabilis strains. Clin. Microbiol. Infect. Suppl. 1:44.
-
- Cieślikowski, T., M. Rzeżnik, and W. Kaca. 2001. Diverse PCR-based fingerprints of several reference P. vulgaris strains correlate with the chemical structure of their O-specific antigens. Clin. Microbiol. Infect. Suppl. 1:3.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

