Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity
- PMID: 12909720
- PMCID: PMC187891
- DOI: 10.1073/pnas.1633803100
Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity
Abstract
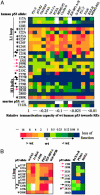
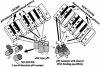
There are many sources of genetic diversity, ranging from programmed mutagenesis in antibody genes to random mutagenesis during species evolution or development of cancer. We propose that mutations in DNA sequence-specific transcription factors that _target response elements (REs) in many genes can also provide for rapid and broad phenotypic diversity, if the mutations lead to altered binding affinities at individual REs. To test this concept, we examined the in vivo transactivation capacity of wild-type human and murine p53 and 25 partial function mutants. The p53s were expressed in yeast from a rheostatable promoter, and the transactivation capacities toward >15 promoter REs upstream of a reporter gene were measured. Surprisingly, there was wide variation in transactivation by the mutant p53s toward the various REs. This is the first study to address directly the impact of mutations in a sequence-specific transcription factor on transactivation from a wide array of REs. We propose a master gene hypothesis for phenotypic diversity where the master gene is a single transcriptional activator (or repressor) that regulates many genes through different REs. Mutations of the master gene can lead to a variety of simultaneous changes in both the selection of _targets and the extent of transcriptional modulation at the individual _targets, resulting in a vast number of potential phenotypes that can be created with minimal mutational changes without altering existing protein-protein interactions.
Figures

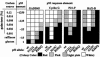
Similar articles
-
Novel human p53 mutations that are toxic to yeast can enhance transactivation of specific promoters and reactivate tumor p53 mutants.Oncogene. 2001 Jun 7;20(26):3409-19. doi: 10.1038/sj.onc.1204457. Oncogene. 2001. PMID: 11423991
-
Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter _target sequence.Mol Cell Biol. 2002 Dec;22(24):8612-25. doi: 10.1128/MCB.22.24.8612-8625.2002. Mol Cell Biol. 2002. PMID: 12446780 Free PMC article.
-
Noncanonical DNA motifs as transactivation _targets by wild type and mutant p53.PLoS Genet. 2008 Jun 27;4(6):e1000104. doi: 10.1371/journal.pgen.1000104. PLoS Genet. 2008. PMID: 18714371 Free PMC article.
-
Potentiating the p53 network.Discov Med. 2010 Jul;10(50):94-100. Discov Med. 2010. PMID: 20670604 Review.
-
Functional diversity in the gene network controlled by the master regulator p53 in humans.Cell Cycle. 2005 Aug;4(8):1026-9. doi: 10.4161/cc.4.8.1904. Epub 2005 Aug 7. Cell Cycle. 2005. PMID: 16082206 Review.
Cited by
-
The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells.Cancer Res. 2012 Aug 15;72(16):3948-57. doi: 10.1158/0008-5472.CAN-11-4134. Epub 2012 Jun 6. Cancer Res. 2012. PMID: 22673234 Free PMC article.
-
Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: results from the BIG 02-98 phase III trial.Breast Cancer Res. 2012 May 2;14(3):R70. doi: 10.1186/bcr3179. Breast Cancer Res. 2012. PMID: 22551440 Free PMC article. Clinical Trial.
-
Characterisation of a novel transcript LNPPS acting as tumour suppressor in bladder cancer via PDCD5-mediated p53 degradation blockage.Clin Transl Med. 2023 Jan;13(1):e1149. doi: 10.1002/ctm2.1149. Clin Transl Med. 2023. PMID: 36578176 Free PMC article.
-
Novel Allosteric Mechanism of Dual p53/MDM2 and p53/MDM4 Inhibition by a Small Molecule.Front Mol Biosci. 2022 Jun 1;9:823195. doi: 10.3389/fmolb.2022.823195. eCollection 2022. Front Mol Biosci. 2022. PMID: 35720128 Free PMC article.
-
∆N-P63α and TA-P63α exhibit intrinsic differences in transactivation specificities that depend on distinct features of DNA _target sites.Onco_target. 2014 Apr 30;5(8):2116-30. doi: 10.18632/onco_target.1845. Onco_target. 2014. PMID: 24926492 Free PMC article.
References
-
- Gearhart, P. J. (2002) Nature 419, 29–31. - PubMed
-
- Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57–70. - PubMed
-
- Lucht, J. M., Mauch-Mani, B., Steiner, H. Y., Metraux, J. P., Ryals, J. & Hohn, B. (2002) Nat. Genet. 30, 311–314. - PubMed
-
- Giraud, A., Radman, M., Matic, I. & Taddei, F. (2001) Curr. Opin. Microbiol. 4, 582–585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

