Ligand-independent activation of estrogen receptor alpha by XBP-1
- PMID: 12954762
- PMCID: PMC203316
- DOI: 10.1093/nar/gkg731
Ligand-independent activation of estrogen receptor alpha by XBP-1
Abstract
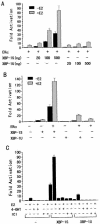
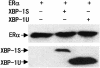
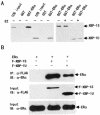
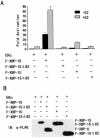
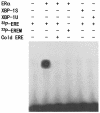
The estrogen receptor (ER) is a member of a large superfamily of nuclear receptors that regulates the transcription of estrogen-responsive genes. Several recent studies have demonstrated that XBP-1 mRNA expression is associated with ERalpha status in breast tumors. However, the role of XBP-1 in ERalpha signaling remains to be elucidated. More recently, two forms of XBP-1 were identified due to its unconventional splicing. We refer to the spliced and unspliced forms of XBP-1 as XBP-1S and XBP-1U, respectively. Here, we report that XBP-1S and XBP-1U enhanced ERalpha-dependent transcriptional activity in a ligand-independent manner. XBP-1S had stronger activity than XBP-1U. The maximal effects of XBP-1S and XBP-1U on ERalpha transactivation were observed when they were co-expressed with full-length ERalpha. SRC-1, the p160 steroid receptor coactivator family member, synergized with XBP-1S or XBP-1U to potentiate ERalpha activity. XBP-1S and XBP-1U bound to the ERalpha both in vitro and in vivo in a ligand-independent fashion. XBP-1S and XBP-1U interacted with the ERalpha region containing the DNA-binding domain. The ERalpha-interacting regions on XBP-1S and XBP-1U have been mapped to two regions, including the N-terminal basic region leucine zipper domain (bZIP) and the C-terminal activation domain. The bZIP-deleted mutants of XBP-1S and XBP-1U completely abolished ERalpha transactivation by XBP-1S and XBP-1U. These findings suggest that XBP-1S and XBP-1U may directly modulate ERalpha signaling in both the absence and presence of estrogen and, therefore, may play important roles in the proliferation of normal and malignant estrogen-regulated tissues.
Figures



Similar articles
-
[XBP-1 enhances the transcriptional activity of estrogen receptor alpha].Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai). 2003 Sep;35(9):829-33. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai). 2003. PMID: 12958656 Chinese.
-
[XBP-1 interacts with estrogen receptor alpha (ERalpha)].Sheng Wu Gong Cheng Xue Bao. 2004 May;20(3):332-6. Sheng Wu Gong Cheng Xue Bao. 2004. PMID: 15971600 Chinese.
-
[Expression of XBP-1 in breast cancer cell lines and its role in ERalpha signaling].Yi Chuan Xue Bao. 2004 Apr;31(4):380-4. Yi Chuan Xue Bao. 2004. PMID: 15487507 Chinese.
-
Molecular action of the estrogen receptor and hormone dependency in breast cancer.Breast Cancer. 2003;10(2):89-96. doi: 10.1007/BF02967632. Breast Cancer. 2003. PMID: 12736560 Review.
-
The physiological role of estrogen receptor functional domains.Essays Biochem. 2021 Dec 17;65(6):867-875. doi: 10.1042/EBC20200167. Essays Biochem. 2021. PMID: 34028522 Free PMC article. Review.
Cited by
-
The Unfolded Protein Response in Breast Cancer.Cancers (Basel). 2018 Sep 21;10(10):344. doi: 10.3390/cancers10100344. Cancers (Basel). 2018. PMID: 30248920 Free PMC article. Review.
-
Master regulators of FGFR2 signalling and breast cancer risk.Nat Commun. 2013;4:2464. doi: 10.1038/ncomms3464. Nat Commun. 2013. PMID: 24043118 Free PMC article.
-
Identification of gene expression signature in estrogen receptor positive breast carcinoma.Biomark Cancer. 2010 Feb 11;2:1-15. doi: 10.4137/BIC.S3793. eCollection 2010. Biomark Cancer. 2010. PMID: 24179381 Free PMC article.
-
NF-κB signaling is required for XBP1 (unspliced and spliced)-mediated effects on antiestrogen responsiveness and cell fate decisions in breast cancer.Mol Cell Biol. 2015 Jan;35(2):379-90. doi: 10.1128/MCB.00847-14. Epub 2014 Nov 3. Mol Cell Biol. 2015. PMID: 25368386 Free PMC article.
-
NCOA3 coactivator is a transcriptional _target of XBP1 and regulates PERK-eIF2α-ATF4 signalling in breast cancer.Oncogene. 2016 Nov 10;35(45):5860-5871. doi: 10.1038/onc.2016.121. Epub 2016 Apr 25. Oncogene. 2016. PMID: 27109102 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

