From gene networks to gene function
- PMID: 14656964
- PMCID: PMC403798
- DOI: 10.1101/gr.1111403
From gene networks to gene function
Abstract
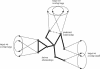
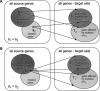
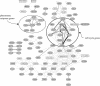
We propose a novel method to identify functionally related genes based on comparisons of neighborhoods in gene networks. This method does not rely on gene sequence or protein structure homologies, and it can be applied to any organism and a wide variety of experimental data sets. The character of the predicted gene relationships depends on the underlying networks;they concern biological processes rather than the molecular function. We used the method to analyze gene networks derived from genome-wide chromatin immunoprecipitation experiments, a large-scale gene deletion study, and from the genomic positions of consensus binding sites for transcription factors of the yeast Saccharomyces cerevisiae. We identified 816 functional relationships between 159 genes and show that these relationships correspond to protein-protein interactions, co-occurrence in the same protein complexes, and/or co-occurrence in abstracts of scientific articles. Our results suggest functions for seven previously uncharacterized yeast genes: KIN3 and YMR269W may be involved in biological processes related to cell growth and/or maintenance, whereas IES6, YEL008W, YEL033W, YHL029C, YMR010W, and YMR031W-A are likely to have metabolic functions.
Figures

Similar articles
-
Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network.Bioessays. 2002 Mar;24(3):267-74. doi: 10.1002/bies.10054. Bioessays. 2002. PMID: 11891763
-
Overexpression of HAM1 gene detoxifies 5-bromodeoxyuridine in the yeast Saccharomyces cerevisiae.Curr Genet. 2007 Nov;52(5-6):203-11. doi: 10.1007/s00294-007-0152-z. Epub 2007 Sep 27. Curr Genet. 2007. PMID: 17899088
-
AVID: an integrative framework for discovering functional relationships among proteins.BMC Bioinformatics. 2005 Jun 1;6:136. doi: 10.1186/1471-2105-6-136. BMC Bioinformatics. 2005. PMID: 15929793 Free PMC article.
-
Transcriptional networks: reverse-engineering gene regulation on a global scale.Curr Opin Microbiol. 2004 Dec;7(6):638-46. doi: 10.1016/j.mib.2004.10.009. Curr Opin Microbiol. 2004. PMID: 15556037 Review.
-
Interaction networks: lessons from large-scale studies in yeast.Proteomics. 2009 Oct;9(20):4799-811. doi: 10.1002/pmic.200900177. Proteomics. 2009. PMID: 19743423 Review.
Cited by
-
Constitutive activation of STAT3 is predictive of poor prognosis in human gastric cancer.J Mol Med (Berl). 2012 Sep;90(9):1037-46. doi: 10.1007/s00109-012-0869-0. Epub 2012 Feb 11. J Mol Med (Berl). 2012. PMID: 22328012
-
How and when should interactome-derived clusters be used to predict functional modules and protein function?Bioinformatics. 2009 Dec 1;25(23):3143-50. doi: 10.1093/bioinformatics/btp551. Epub 2009 Sep 21. Bioinformatics. 2009. PMID: 19770263 Free PMC article.
-
Large-scale analyses identify a cluster of novel long noncoding RNAs as potential competitive endogenous RNAs in progression of hepatocellular carcinoma.Aging (Albany NY). 2019 Nov 23;11(22):10422-10453. doi: 10.18632/aging.102468. Epub 2019 Nov 23. Aging (Albany NY). 2019. PMID: 31761783 Free PMC article.
-
Mutual information estimation reveals global associations between stimuli and biological processes.BMC Bioinformatics. 2009 Jan 30;10 Suppl 1(Suppl 1):S52. doi: 10.1186/1471-2105-10-S1-S52. BMC Bioinformatics. 2009. PMID: 19208155 Free PMC article.
-
Identification of gene expression profile in the rat brain resulting from acute alcohol intoxication.Mol Biol Rep. 2014 Dec;41(12):8303-17. doi: 10.1007/s11033-014-3731-3. Epub 2014 Sep 14. Mol Biol Rep. 2014. Retraction in: Mol Biol Rep. 2015 Jul;42(7):1241. doi: 10.1007/s11033-015-3873-y PMID: 25218841 Retracted.
References
-
- Alepuz, P.M., Cunningham, K.W., and Estruch, F. 1997. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 26: 91-98. - PubMed
-
- Bader, G.D. and Hogue, C.W. 2002. Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol. 20: 991-997. - PubMed
-
- Blake, J. and Harris, M. 2003. The Gene Ontology (GO) Project: Structured vocabularies for molecular biology and their application to genome and expression analysis. In Current protocols in bioinformatics.(eds. A. Baxevanis, et al.), J. Wiley, New York. - PubMed
WEB SITE REFERENCES
-
- http://db.yeastgenome.org/cgi-bin/SGD/GO/goTermMapper; goTermMapper from SGD.
-
- http://srs.ebi.ac.uk; SRS server.
-
- http://www.ebi.ac.uk/proteome/; EBI Proteome Analysis Database.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
