The prokaryotic selenoproteome
- PMID: 15105824
- PMCID: PMC1299047
- DOI: 10.1038/sj.embor.7400126
The prokaryotic selenoproteome
Abstract
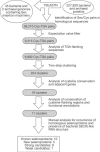
In the genetic code, the UGA codon has a dual function as it encodes selenocysteine (Sec) and serves as a stop signal. However, only the translation terminator function is used in gene annotation programs, resulting in misannotation of selenoprotein genes. Here, we applied two independent bioinformatics approaches to characterize a selenoprotein set in prokaryotic genomes. One method searched for selenoprotein genes by identifying RNA stem-loop structures, selenocysteine insertion sequence elements; the second approach identified Sec/Cys pairs in homologous sequences. These analyses identified all or almost all selenoproteins in completely sequenced bacterial and archaeal genomes and provided a view on the distribution and composition of prokaryotic selenoproteomes. In addition, lineage-specific and core selenoproteins were detected, which provided insights into the mechanisms of selenoprotein evolution. Characterization of selenoproteomes allows interpretation of other UGA codons in completed genomes of prokaryotes as terminators, addressing the UGA dual-function problem.
Figures
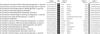

Similar articles
-
Characterization of mammalian selenoproteomes.Science. 2003 May 30;300(5624):1439-43. doi: 10.1126/science.1083516. Science. 2003. PMID: 12775843
-
Nematode selenoproteome: the use of the selenocysteine insertion system to decode one codon in an animal genome?Nucleic Acids Res. 2005 Apr 20;33(7):2227-38. doi: 10.1093/nar/gki507. Print 2005. Nucleic Acids Res. 2005. PMID: 15843685 Free PMC article.
-
Asgard archaeal selenoproteome reveals a roadmap for the archaea-to-eukaryote transition of selenocysteine incorporation machinery.ISME J. 2024 Jan 8;18(1):wrae111. doi: 10.1093/ismejo/wrae111. ISME J. 2024. PMID: 38896033 Free PMC article.
-
Selenoprotein synthesis in archaea.Biofactors. 2001;14(1-4):75-83. doi: 10.1002/biof.5520140111. Biofactors. 2001. PMID: 11568443 Review.
-
Bioinformatics of Selenoproteins.Antioxid Redox Signal. 2020 Sep 1;33(7):525-536. doi: 10.1089/ars.2020.8044. Epub 2020 Apr 23. Antioxid Redox Signal. 2020. PMID: 32031018 Free PMC article. Review.
Cited by
-
Role of Selenoproteins in Bacterial Pathogenesis.Biol Trace Elem Res. 2019 Nov;192(1):69-82. doi: 10.1007/s12011-019-01877-2. Epub 2019 Sep 5. Biol Trace Elem Res. 2019. PMID: 31489516 Free PMC article. Review.
-
A method for identification of selenoprotein genes in archaeal genomes.Genomics Proteomics Bioinformatics. 2009 Jun;7(1-2):62-70. doi: 10.1016/S1672-0229(08)60034-0. Genomics Proteomics Bioinformatics. 2009. PMID: 19591793 Free PMC article.
-
Genetic analysis of selenocysteine biosynthesis in the archaeon Methanococcus maripaludis.Mol Microbiol. 2011 Jul;81(1):249-58. doi: 10.1111/j.1365-2958.2011.07690.x. Epub 2011 May 18. Mol Microbiol. 2011. PMID: 21564332 Free PMC article.
-
Evolution of the Selenoproteome in Helicobacter pylori and Epsilonproteobacteria.Genome Biol Evol. 2015 Sep 4;7(9):2692-704. doi: 10.1093/gbe/evv177. Genome Biol Evol. 2015. PMID: 26342139 Free PMC article.
-
Selenocysteine insertion directed by the 3'-UTR SECIS element in Escherichia coli.Nucleic Acids Res. 2005 Apr 29;33(8):2486-92. doi: 10.1093/nar/gki547. Print 2005. Nucleic Acids Res. 2005. PMID: 15863725 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353: 273–276 - PubMed
-
- Böck A (2000) Biosynthesis of selenoproteins—an overview. Biofactors 11: 77–78 - PubMed
-
- Dsouza M, Larsen N, Overbeek R (1997) Searching for patterns in genomic data. Trends Genet 13: 497–498 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

