Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes
- PMID: 15613353
- PMCID: PMC538569
- DOI: 10.1128/JVI.79.2.1262-1270.2005
Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes
Abstract
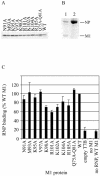
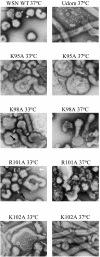
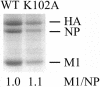
Several functions required for the replication of influenza A viruses have been attributed to the viral matrix protein (M1), and a number of studies have focused on a region of the M1 protein designated "helix six." This region contains an exposed positively charged stretch of amino acids, including the motif 101-RKLKR-105, which has been identified as a nuclear localization signal, but several studies suggest that this domain is also involved in functions such as binding to the ribonucleoprotein genome segments (RNPs), membrane association, interaction with the viral nuclear export protein, and virus assembly. In order to define M1 functions in more detail, a series of mutants containing alanine substitutions in the helix six region were generated in A/WSN/33 virus. These were analyzed for RNP-binding function, their capacity to incorporate into infectious viruses by using reverse genetics, the replication properties of rescued viruses, and the morphological phenotypes of the mutant virus particles. The most notable effect that was identified concerned single amino acid substitution mutants that caused significant alterations to the morphology of budded viruses. Whereas A/WSN/33 virus generally forms particles that are predominantly spherical, observations made by negative stain electron microscopy showed that several of the mutant virions, such as K95A, K98A, R101A, and K102A, display a wide range of shapes and sizes that varied in a temperature-dependent manner. The K102A mutant is particularly interesting in that it can form extended filamentous particles. These results support the proposition that the helix six domain is involved in the process of virus assembly.
Figures




Similar articles
-
Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs.J Virol. 2003 Jun;77(12):7078-92. doi: 10.1128/jvi.77.12.7078-7092.2003. J Virol. 2003. Retraction in: J Virol. 2006 Oct;80(20):10289. doi: 10.1128/JVI.01632-06 PMID: 12768027 Free PMC article. Retracted.
-
Introduction of a temperature-sensitive phenotype into influenza A/WSN/33 virus by altering the basic amino acid domain of influenza virus matrix protein.J Virol. 2004 Sep;78(18):9585-91. doi: 10.1128/JVI.78.18.9585-9591.2004. J Virol. 2004. PMID: 15331690 Free PMC article.
-
Reverse genetics studies on the filamentous morphology of influenza A virus.J Gen Virol. 2003 Mar;84(Pt 3):517-527. doi: 10.1099/vir.0.18803-0. J Gen Virol. 2003. PMID: 12604801
-
Structure of influenza virus ribonucleoprotein complexes and their packaging into virions.Rev Med Virol. 2010 Nov;20(6):380-91. doi: 10.1002/rmv.666. Rev Med Virol. 2010. PMID: 20853340 Free PMC article. Review.
-
[Structure, function and regulation of expression of influenza virus matrix M1 protein].Nihon Rinsho. 1997 Oct;55(10):2581-6. Nihon Rinsho. 1997. PMID: 9360375 Review. Japanese.
Cited by
-
Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer.PLoS Biol. 2020 Sep 30;18(9):e3000827. doi: 10.1371/journal.pbio.3000827. eCollection 2020 Sep. PLoS Biol. 2020. PMID: 32997652 Free PMC article.
-
Structural Analysis of the Roles of Influenza A Virus Membrane-Associated Proteins in Assembly and Morphology.J Virol. 2015 Sep;89(17):8957-66. doi: 10.1128/JVI.00592-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085153 Free PMC article.
-
Influenza A: understanding the viral life cycle.Yale J Biol Med. 2009 Dec;82(4):153-9. Yale J Biol Med. 2009. PMID: 20027280 Free PMC article. Review.
-
Specific nucleoprotein residues affect influenza virus morphology.J Virol. 2014 Feb;88(4):2227-34. doi: 10.1128/JVI.03354-13. Epub 2013 Dec 11. J Virol. 2014. PMID: 24335312 Free PMC article.
-
An unbiased genetic screen reveals the polygenic nature of the influenza virus anti-interferon response.J Virol. 2014 May;88(9):4632-46. doi: 10.1128/JVI.00014-14. Epub 2014 Feb 26. J Virol. 2014. PMID: 24574395 Free PMC article.
References
-
- Arzt, S., F. Baudin, A. Barge, P. Timmins, W. P. Burmeister, and R. W. Ruigrok. 2001. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology 279:439-446. - PubMed
-
- Barman, S., A. Ali, E. K. Hui, L. Adhikary, and D. P. Nayak. 2001. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 77:61-69. - PubMed
-
- Baudin, F., I. Petit, W. Weissenhorn, and R. W. Ruigrok. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102-108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

