SIRT1 regulates HIV transcription via Tat deacetylation
- PMID: 15719057
- PMCID: PMC546329
- DOI: 10.1371/journal.pbio.0030041
SIRT1 regulates HIV transcription via Tat deacetylation
Abstract
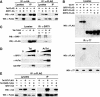
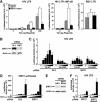
The human immunodeficiency virus (HIV) Tat protein is acetylated by the transcriptional coactivator p300, a necessary step in Tat-mediated transactivation. We report here that Tat is deacetylated by human sirtuin 1 (SIRT1), a nicotinamide adenine dinucleotide-dependent class III protein deacetylase in vitro and in vivo. Tat and SIRT1 coimmunoprecipitate and synergistically activate the HIV promoter. Conversely, knockdown of SIRT1 via small interfering RNAs or treatment with a novel small molecule inhibitor of the SIRT1 deacetylase activity inhibit Tat-mediated transactivation of the HIV long terminal repeat. Tat transactivation is defective in SIRT1-null mouse embryonic fibroblasts and can be rescued by expression of SIRT1. These results support a model in which cycles of Tat acetylation and deacetylation regulate HIV transcription. SIRT1 recycles Tat to its unacetylated form and acts as a transcriptional coactivator during Tat transactivation.
Figures

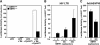

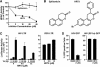
Similar articles
-
Tat acetylation: a regulatory switch between early and late phases in HIV transcription elongation.Novartis Found Symp. 2004;259:182-93; discussion 193-6, 223-5. Novartis Found Symp. 2004. PMID: 15171254 Review.
-
Resveratrol inhibited Tat-induced HIV-1 LTR transactivation via NAD(+)-dependent SIRT1 activity.Life Sci. 2009 Sep 23;85(13-14):484-9. doi: 10.1016/j.lfs.2009.07.014. Epub 2009 Aug 5. Life Sci. 2009. PMID: 19664641
-
SIRT1 regulates Tat-induced HIV-1 transactivation through activating AMP-activated protein kinase.Virus Res. 2009 Dec;146(1-2):51-7. doi: 10.1016/j.virusres.2009.08.005. Epub 2009 Aug 29. Virus Res. 2009. PMID: 19720090
-
Cloning, chromosomal characterization and mapping of the NAD-dependent histone deacetylases gene sirtuin 1.Int J Mol Med. 2006 Jan;17(1):59-67. Int J Mol Med. 2006. PMID: 16328012
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
Cited by
-
Epigenetics and Genetics of Viral Latency.Cell Host Microbe. 2016 May 11;19(5):619-28. doi: 10.1016/j.chom.2016.04.008. Cell Host Microbe. 2016. PMID: 27173930 Free PMC article. Review.
-
The sirtuin family in health and disease.Signal Transduct _target Ther. 2022 Dec 29;7(1):402. doi: 10.1038/s41392-022-01257-8. Signal Transduct _target Ther. 2022. PMID: 36581622 Free PMC article. Review.
-
An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer.PLoS Biol. 2007 Jan;5(1):e9. doi: 10.1371/journal.pbio.0050009. PLoS Biol. 2007. PMID: 17194214 Free PMC article.
-
miR-224 suppresses HBV replication posttranscriptionally through inhibiting SIRT1-mediated autophagy.Int J Clin Exp Pathol. 2018 Jan 1;11(1):189-198. eCollection 2018. Int J Clin Exp Pathol. 2018. PMID: 31938100 Free PMC article.
-
Transcription Regulation by Class III Histone Deacetylases (HDACs)-Sirtuins.Transl Oncogenomics. 2008 Apr 23;3:53-65. doi: 10.4137/tog.s483. Transl Oncogenomics. 2008. PMID: 21566744 Free PMC article.
References
-
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. - PubMed
-
- Toohey MG, Jones KA. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes Dev. 1989;3:265–282. - PubMed
-
- Rosen CA, Sodroski JG, Haseltine WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. - PubMed
-
- Feng S, Holland EC. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. - PubMed
-
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

