Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein
- PMID: 15755749
- PMCID: PMC1062877
- DOI: 10.1093/nar/gki292
Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein
Abstract
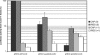
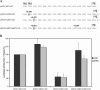
The hepatitis C virus (HCV) genome contains an internal ribosome entry site (IRES) followed by a large open reading frame coding for a polyprotein that is cleaved into 10 proteins. An additional HCV protein, the F protein, was recently suggested to result from a +1 frameshift by a minority of ribosomes that initiated translation at the HCV AUG initiator codon of the polyprotein. In the present study, we reassessed the mechanism accounting for the synthesis of the F protein by measuring the expression in cultured cells of a luciferase reporter gene with an insertion encompassing the IRES plus the beginning of the HCV-coding region preceding the luciferase-coding sequence. The insertion was such that luciferase expression was either in the +1 reading frame relative to the HCV AUG initiator codon, mimicking the expression of the F protein, or in-frame with this AUG, mimicking the expression of the polyprotein. Introduction of a stop codon at various positions in-frame with the AUG initiator codon and substitution of this AUG with UAC inhibited luciferase expression in the 0 reading frame but not in the +1 reading frame, ruling out that the synthesis of the F protein results from a +1 frameshift. Introduction of a stop codon at various positions in the +1 reading frame identified the codon overlapping codon 26 of the polyprotein in the +1 reading frame as the translation start site for the F protein. This codon 26(+1) is either GUG or GCG in the viral variants. Expression of the F protein strongly increased when codon 26(+1) was replaced with AUG, or when its context was mutated into an optimal Kozak context, but was severely decreased in the presence of low concentrations of edeine. These observations are consistent with a Met-tRNA(i)-dependent initiation of translation at a non-AUG codon for the synthesis of the F protein.
Figures
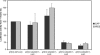
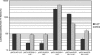
Similar articles
-
Internal translation initiation stimulates expression of the ARF/core+1 open reading frame of HCV genotype 1b.Virus Res. 2011 Jan;155(1):213-20. doi: 10.1016/j.virusres.2010.10.007. Epub 2010 Oct 17. Virus Res. 2011. PMID: 20959129
-
Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA.Virology. 1996 Aug 1;222(1):31-42. doi: 10.1006/viro.1996.0395. Virology. 1996. PMID: 8806485
-
Expression studies of the HCV-1a core+1 open reading frame in mammalian cells.Virus Res. 2008 May;133(2):123-35. doi: 10.1016/j.virusres.2007.10.019. Epub 2008 Feb 19. Virus Res. 2008. PMID: 18243391
-
The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others.Semin Liver Dis. 2005 Feb;25(1):105-17. doi: 10.1055/s-2005-864786. Semin Liver Dis. 2005. PMID: 15732002 Review.
-
tRNA-mimicry in IRES-mediated translation and recoding.RNA Biol. 2016 Nov;13(11):1068-1074. doi: 10.1080/15476286.2016.1219833. Epub 2016 Aug 11. RNA Biol. 2016. PMID: 27654067 Free PMC article. Review.
Cited by
-
Molecular evolution of the genomic RNA of Apple stem grooving capillovirus.J Mol Evol. 2012 Oct;75(3-4):92-101. doi: 10.1007/s00239-012-9518-z. Epub 2012 Nov 13. J Mol Evol. 2012. PMID: 23149596
-
The evolution of genome compression and genomic novelty in RNA viruses.Genome Res. 2007 Oct;17(10):1496-504. doi: 10.1101/gr.6305707. Epub 2007 Sep 4. Genome Res. 2007. PMID: 17785537 Free PMC article.
-
A Novel Cis-Acting RNA Structural Element Embedded in the Core Coding Region of the Hepatitis C Virus Genome Directs Internal Translation Initiation of the Overlapping Core+1 ORF.Int J Mol Sci. 2020 Sep 22;21(18):6974. doi: 10.3390/ijms21186974. Int J Mol Sci. 2020. PMID: 32972019 Free PMC article.
-
HELZ2: a new, interferon-regulated, human 3'-5' exoribonuclease of the RNB family is expressed from a non-canonical initiation codon.Nucleic Acids Res. 2023 Sep 22;51(17):9279-9293. doi: 10.1093/nar/gkad673. Nucleic Acids Res. 2023. PMID: 37602378 Free PMC article.
-
Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death.Virology. 2008 Jul 5;376(2):397-407. doi: 10.1016/j.virol.2008.03.027. Epub 2008 May 2. Virology. 2008. PMID: 18455749 Free PMC article.
References
-
- Lefkowitch J.H., Schiff E.R., Davis G.L., Perrillo R.P., Lindsay K., Bodenheimer H.C., Balart L.A., Ortego T.J., Payne J., Dienstag J.L. Pathological diagnosis of chronic hepatitis C: a multicenter comparative study with chronic hepatitis B. The Hepatitis Interventional Therapy Group. Gastroenterology. 1993;104:595–603. - PubMed
-
- Scheuer P.J., Ashrafzadeh P., Sherlock S., Brown D., Dusheiko G.M. The pathology of hepatitis C. Hepatology. 1992;15:567–571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

