Exercise responsive genes measured in peripheral blood of women with chronic fatigue syndrome and matched control subjects
- PMID: 15790422
- PMCID: PMC1079885
- DOI: 10.1186/1472-6793-5-5
Exercise responsive genes measured in peripheral blood of women with chronic fatigue syndrome and matched control subjects
Abstract
Background: Chronic fatigue syndrome (CFS) is defined by debilitating fatigue that is exacerbated by physical or mental exertion. To search for markers of CFS-associated post-exertional fatigue, we measured peripheral blood gene expression profiles of women with CFS and matched controls before and after exercise challenge.
Results: Women with CFS and healthy, age-matched, sedentary controls were exercised on a stationary bicycle at 70% of their predicted maximum workload. Blood was obtained before and after the challenge, total RNA was extracted from mononuclear cells, and signal intensity of the labeled cDNA hybridized to a 3800-gene oligonucleotide microarray was measured. We identified differences in gene expression among and between subject groups before and after exercise challenge and evaluated differences in terms of Gene Ontology categories. Exercise-responsive genes differed between CFS patients and controls. These were in genes classified in chromatin and nucleosome assembly, cytoplasmic vesicles, membrane transport, and G protein-coupled receptor ontologies. Differences in ion transport and ion channel activity were evident at baseline and were exaggerated after exercise, as evidenced by greater numbers of differentially expressed genes in these molecular functions.
Conclusion: These results highlight the potential use of an exercise challenge combined with microarray gene expression analysis in identifying gene ontologies associated with CFS.
Figures
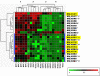
 are classified to the GO category of binding (Molecular Function); and/or # are classified in the GO category of metabolism (Biological Process). The subjects group into 4 clusters which approximate to: 1) Control subjects before exercise (Con0); 2) CFS cases before exercise (CFS0); 3) Control subjects after exercise (Con24); and 4) CFS cases after exercise (CFS24).
are classified to the GO category of binding (Molecular Function); and/or # are classified in the GO category of metabolism (Biological Process). The subjects group into 4 clusters which approximate to: 1) Control subjects before exercise (Con0); 2) CFS cases before exercise (CFS0); 3) Control subjects after exercise (Con24); and 4) CFS cases after exercise (CFS24).
Similar articles
-
Utility of the blood for gene expression profiling and biomarker discovery in chronic fatigue syndrome.Dis Markers. 2002;18(4):193-9. doi: 10.1155/2002/892374. Dis Markers. 2002. PMID: 12590173 Free PMC article.
-
Gene expression profile exploration of a large dataset on chronic fatigue syndrome.Pharmacogenomics. 2006 Apr;7(3):429-40. doi: 10.2217/14622416.7.3.429. Pharmacogenomics. 2006. PMID: 16610953
-
Identification of marker genes for differential diagnosis of chronic fatigue syndrome.Mol Med. 2008 Sep-Oct;14(9-10):599-607. doi: 10.2119/2007-00059.Saiki. Mol Med. 2008. PMID: 18596870 Free PMC article.
-
Whole blood gene expression in adolescent chronic fatigue syndrome: an exploratory cross-sectional study suggesting altered B cell differentiation and survival.J Transl Med. 2017 May 11;15(1):102. doi: 10.1186/s12967-017-1201-0. J Transl Med. 2017. PMID: 28494812 Free PMC article.
-
[Identification and application of marker genes for differential diagnosis of chronic fatigue syndrome].Nihon Rinsho. 2007 Jun;65(6):1029-33. Nihon Rinsho. 2007. PMID: 17561693 Review. Japanese.
Cited by
-
The Pathobiology of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: The Case for Neuroglial Failure.Front Cell Neurosci. 2022 May 9;16:888232. doi: 10.3389/fncel.2022.888232. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35614970 Free PMC article.
-
A double-blind, placebo-controlled, randomized, clinical trial of the TLR-3 agonist rintatolimod in severe cases of chronic fatigue syndrome.PLoS One. 2012;7(3):e31334. doi: 10.1371/journal.pone.0031334. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22431963 Free PMC article. Clinical Trial.
-
A class comparison method with filtering-enhanced variable selection for high-dimensional data sets.Stat Med. 2008 Dec 10;27(28):5834-49. doi: 10.1002/sim.3405. Stat Med. 2008. PMID: 18781559 Free PMC article.
-
Post-Exertional Malaise Is Associated with Hypermetabolism, Hypoacetylation and Purine Metabolism Deregulation in ME/CFS Cases.Diagnostics (Basel). 2019 Jul 4;9(3):70. doi: 10.3390/diagnostics9030070. Diagnostics (Basel). 2019. PMID: 31277442 Free PMC article.
-
The status of and future research into Myalgic Encephalomyelitis and Chronic Fatigue Syndrome: the need of accurate diagnosis, objective assessment, and acknowledging biological and clinical subgroups.Front Physiol. 2014 Mar 27;5:109. doi: 10.3389/fphys.2014.00109. eCollection 2014. Front Physiol. 2014. PMID: 24734022 Free PMC article.
References
-
- Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann Intern Med. 1994;121:953–959. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

