Specificity and mechanism of the histone methyltransferase Pr-Set7
- PMID: 15933069
- PMCID: PMC1151661
- DOI: 10.1101/gad.1315905
Specificity and mechanism of the histone methyltransferase Pr-Set7
Abstract
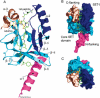
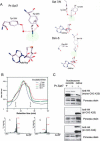
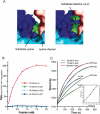
Methylation of lysine residues of histones is an important epigenetic mark that correlates with functionally distinct regions of chromatin. We present here the crystal structure of a ternary complex of the enzyme Pr-Set7 (also known as Set8) that methylates Lys 20 of histone H4 (H4-K20). We show that the enzyme is exclusively a mono-methylase and is therefore responsible for a signaling role quite distinct from that established by other enzymes that _target this histone residue. We provide evidence from NMR for the C-flanking domains of SET proteins becoming ordered upon addition of AdoMet cofactor and develop a model for the catalytic cycle of these enzymes. The crystal structure reveals the basis of the specificity of the enzyme for H4-K20 because a histidine residue within the substrate, close to the _target lysine, is required for completion of the active site. We also show how a highly variable component of the SET domain is responsible for many of the enzymes' interactions with its _target histone peptide and probably also how this part of the structure ensures that Pr-Set7 is nucleosome specific.
Figures


Similar articles
-
Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase.Genes Dev. 2005 Jun 15;19(12):1455-65. doi: 10.1101/gad.1318405. Epub 2005 Jun 2. Genes Dev. 2005. PMID: 15933070 Free PMC article.
-
Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase.Curr Biol. 2002 Jul 9;12(13):1086-99. doi: 10.1016/s0960-9822(02)00924-7. Curr Biol. 2002. PMID: 12121615
-
Product specificity and mechanism of protein lysine methyltransferases: insights from the histone lysine methyltransferase SET8.Biochemistry. 2008 Jun 24;47(25):6671-7. doi: 10.1021/bi800244s. Biochemistry. 2008. PMID: 18512960
-
SET domain protein lysine methyltransferases: Structure, specificity and catalysis.Cell Mol Life Sci. 2006 Dec;63(23):2755-63. doi: 10.1007/s00018-006-6274-5. Cell Mol Life Sci. 2006. PMID: 17013555 Free PMC article. Review.
-
[The biological functions of lysine methyltransferase PR-SET7].Yi Chuan. 2013 Mar;35(3):241-54. doi: 10.3724/sp.j.1005.2013.00241. Yi Chuan. 2013. PMID: 23575530 Review. Chinese.
Cited by
-
PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.Nat Struct Mol Biol. 2012 Sep;19(9):916-24. doi: 10.1038/nsmb.2353. Epub 2012 Aug 5. Nat Struct Mol Biol. 2012. PMID: 22864287 Free PMC article.
-
Interaction of SET domains with histones and nucleic acid structures in active chromatin.Clin Epigenetics. 2011 Apr;2(1):17-25. doi: 10.1007/s13148-010-0015-1. Epub 2011 Jan 14. Clin Epigenetics. 2011. PMID: 22704267 Free PMC article.
-
Structural dynamics of protein lysine methylation and demethylation.Mutat Res. 2007 May 1;618(1-2):102-15. doi: 10.1016/j.mrfmmm.2006.05.041. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17374386 Free PMC article. Review.
-
Comparative analyses of SUV420H1 isoforms and SUV420H2 reveal differences in their cellular localization and effects on myogenic differentiation.PLoS One. 2010 Dec 29;5(12):e14447. doi: 10.1371/journal.pone.0014447. PLoS One. 2010. PMID: 21206904 Free PMC article.
-
Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations.PLoS One. 2013 Dec 19;8(12):e83737. doi: 10.1371/journal.pone.0083737. eCollection 2013. PLoS One. 2013. PMID: 24367611 Free PMC article.
References
-
- Breiling A. and Orlando, V. 2002. SET domain proteins reSET gene expression. Nat. Struct. Biol. 9: 894-896. - PubMed
-
- Chuikov S., Kurash, J.K., Wilson, J.R., Xiao, B., Justin, N., Ivanov, G.S., McKinney, K., Tempst, P., Prives, C., Gamblin, S.J., et al. 2004. Regulation of p53 activity through lysine methylation. Nature 432: 353-360. - PubMed
-
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760-763. - PubMed
-
- Davey C.A., Sargent, D.F., Luger, K., Maeder, A.W., and Richmond, T.J. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319: 1097-1113. - PubMed
-
- Fang J., Feng, Q., Ketel, C.S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J.A., and Zhang, Y. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12: 1086-1099. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
