Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation
- PMID: 15961999
- PMCID: PMC1173159
- DOI: 10.1038/sj.emboj.7600719
Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation
Abstract
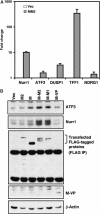
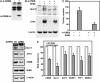
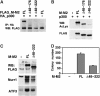
Salivary gland tumors, a group of histologically diverse benign and malignant neoplasms, represent a challenging problem for diagnosis and treatment. A specific recurring t(11;19)(q21;p13) translocation is associated with two types of salivary gland tumors, mucoepidermoid carcinomas and Warthin's tumors. This translocation generates a fusion protein comprised of the N-terminal CREB (cAMP response element-binding protein)-binding domain of the CREB regulator MECT1 (Mucoepidermoid carcinoma translocated-1) and the C-terminal transcriptional activation domain of the Notch coactivator Mastermind-like 2 (MAML2). Here, we demonstrate that the MECT1-MAML2 fusion protein induces expression of multiple genes known to be CREB transcriptional _targets. MECT1-MAML2 was found to bind to CREB, recruit p300/CBP into the CREB complex through a binding domain on MAML2, and constitutively activate CREB-dependent transcription. The transforming activity of MECT1-MAML2 was markedly reduced by blocking CREB DNA binding. Thus, this fusion oncogene mimics constitutive activation of cAMP signaling, by activating CREB directly. This study has identified a novel, critical mechanism of transformation for an oncogene associated very specifically with salivary gland tumors, and identified potential _targets for the development of novel therapies.
Figures




Similar articles
-
Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes.Cancer Res. 2005 Aug 15;65(16):7137-44. doi: 10.1158/0008-5472.CAN-05-1125. Cancer Res. 2005. PMID: 16103063
-
CRTC1/MAML2 gain-of-function interactions with MYC create a gene signature predictive of cancers with CREB-MYC involvement.Proc Natl Acad Sci U S A. 2014 Aug 12;111(32):E3260-8. doi: 10.1073/pnas.1319176111. Epub 2014 Jul 28. Proc Natl Acad Sci U S A. 2014. PMID: 25071166 Free PMC article.
-
Molecular Basis for the Mechanism of Constitutive CBP/p300 Coactivator Recruitment by CRTC1-MAML2 and Its Implications in cAMP Signaling.Biochemistry. 2015 Sep 8;54(35):5439-46. doi: 10.1021/acs.biochem.5b00332. Epub 2015 Aug 21. Biochemistry. 2015. PMID: 26274502 Free PMC article.
-
t(11;19) translocation and CRTC1-MAML2 fusion oncogene in mucoepidermoid carcinoma.Oral Oncol. 2009 Jan;45(1):2-9. doi: 10.1016/j.oraloncology.2008.03.012. Epub 2008 May 16. Oral Oncol. 2009. PMID: 18486532 Review.
-
Clinicopathologic and genetic features of primary bronchopulmonary mucoepidermoid carcinoma: the MD Anderson Cancer Center experience and comprehensive review of the literature.Virchows Arch. 2017 Jun;470(6):619-626. doi: 10.1007/s00428-017-2104-4. Epub 2017 Mar 25. Virchows Arch. 2017. PMID: 28343305 Review.
Cited by
-
Salivary gland cancer stem cells.Oral Oncol. 2013 Sep;49(9):845-853. doi: 10.1016/j.oraloncology.2013.05.013. Epub 2013 Jun 28. Oral Oncol. 2013. PMID: 23810400 Free PMC article. Review.
-
Dysregulated gene subnetworks in breast invasive carcinoma reveal novel tumor suppressor genes.Sci Rep. 2024 Jul 8;14(1):15691. doi: 10.1038/s41598-024-59953-0. Sci Rep. 2024. PMID: 38977697 Free PMC article.
-
Inhibition of notch signaling blocks growth of glioblastoma cell lines and tumor neurospheres.Genes Cancer. 2010 Aug;1(8):822-35. doi: 10.1177/1947601910383564. Genes Cancer. 2010. PMID: 21127729 Free PMC article.
-
Dysregulated CRTC1 activity is a novel component of PGE2 signaling that contributes to colon cancer growth.Oncogene. 2016 May 19;35(20):2602-14. doi: 10.1038/onc.2015.283. Epub 2015 Aug 24. Oncogene. 2016. PMID: 26300003
-
Prognostic impact of CRTC1/3-MAML2 fusions in salivary gland mucoepidermoid carcinoma: A multiinstitutional retrospective study.Cancer Sci. 2020 Nov;111(11):4195-4204. doi: 10.1111/cas.14632. Epub 2020 Sep 14. Cancer Sci. 2020. PMID: 32860299 Free PMC article.
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 - PubMed
-
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M (2004) Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol 14: 2156–2161 - PubMed
-
- Borrow J, Stanton VP Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dube I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE (1996) The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 14: 33–41 - PubMed
-
- Carlesso N, Aster JC, Sklar J, Scadden DT (1999) Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood 93: 838–848 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

