Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma
- PMID: 16163384
- PMCID: PMC1276176
- DOI: 10.1038/sj.emboj.7600819
Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma
Abstract
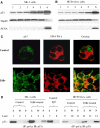
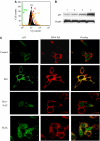
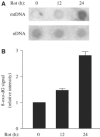
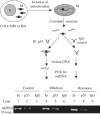
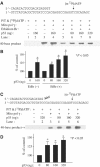
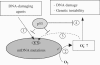
Mitochondrial DNA (mtDNA) mutations and deletions are frequently observed in cancer, and contribute to altered energy metabolism, increased reactive oxygen species (ROS), and attenuated apoptotic response to anticancer agents. The mechanisms by which cells maintain mitochondrial genomic integrity and the reason why cancer cells exhibit more frequent mtDNA mutations remain unclear. Here, we report that the tumor suppressor molecule p53 has a novel role in maintaining mitochondrial genetic stability through its ability to translocate to mitochondria and interact with mtDNA polymerase gamma (pol gamma) in response to mtDNA damage induced by exogenous and endogenous insults including ROS. The p53 protein physically interacts with mtDNA and pol gamma, and enhances the DNA replication function of pol gamma. Loss of p53 results in a significant increase in mtDNA vulnerability to damage, leading to increased frequency of in vivo mtDNA mutations, which are reversed by stable transfection of wild-type p53. This study provides a mechanistic explanation for the accelerating genetic instability and increased ROS stress in cancer cells associated with loss of p53.
Figures



Similar articles
-
Exercise-induced mitochondrial p53 repairs mtDNA mutations in mutator mice.Skelet Muscle. 2016 Jan 31;6:7. doi: 10.1186/s13395-016-0075-9. eCollection 2016. Skelet Muscle. 2016. Retraction in: Skelet Muscle. 2021 Mar 30;11(1):8. doi: 10.1186/s13395-021-00264-7 PMID: 26834962 Free PMC article. Retracted.
-
RECQL4 and p53 potentiate the activity of polymerase γ and maintain the integrity of the human mitochondrial genome.Carcinogenesis. 2014 Jan;35(1):34-45. doi: 10.1093/carcin/bgt315. Epub 2013 Sep 25. Carcinogenesis. 2014. PMID: 24067899
-
Mitochondrial DNA damage and its consequences for mitochondrial gene expression.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):979-91. doi: 10.1016/j.bbagrm.2012.06.002. Epub 2012 Jun 19. Biochim Biophys Acta. 2012. PMID: 22728831 Free PMC article. Review.
-
Mitochondrial DNA maintenance is regulated in human hepatoma cells by glycogen synthase kinase 3β and p53 in response to tumor necrosis factor α.PLoS One. 2012;7(7):e40879. doi: 10.1371/journal.pone.0040879. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911714 Free PMC article.
-
p53 as guardian of the mitochondrial genome.FEBS Lett. 2016 Apr;590(7):924-34. doi: 10.1002/1873-3468.12061. Epub 2016 Feb 3. FEBS Lett. 2016. PMID: 26780878 Free PMC article. Review.
Cited by
-
Part III. Molecular changes induced by high nitric oxide adaptation in human breast cancer cell line BT-20 (BT-20-HNO): a switch from aerobic to anaerobic metabolism.Tumour Biol. 2013 Feb;34(1):403-13. doi: 10.1007/s13277-012-0564-3. Epub 2012 Dec 14. Tumour Biol. 2013. PMID: 23238817
-
Loss of p53 in stromal fibroblasts promotes epithelial cell invasion through redox-mediated ICAM1 signal.Free Radic Biol Med. 2013 May;58:1-13. doi: 10.1016/j.freeradbiomed.2013.01.011. Epub 2013 Jan 29. Free Radic Biol Med. 2013. PMID: 23376231 Free PMC article.
-
Amelioration of premature aging in mtDNA mutator mouse by exercise: the interplay of oxidative stress, PGC-1α, p53, and DNA damage. A hypothesis.Curr Opin Genet Dev. 2016 Jun;38:127-132. doi: 10.1016/j.gde.2016.06.011. Epub 2016 Aug 3. Curr Opin Genet Dev. 2016. PMID: 27497229 Free PMC article. Review.
-
Investigation of the association between mitochondrial DNA and p53 gene mutations in transitional cell carcinoma of the bladder.Oncol Lett. 2016 Oct;12(4):2872-2879. doi: 10.3892/ol.2016.5000. Epub 2016 Aug 11. Oncol Lett. 2016. PMID: 27698873 Free PMC article.
-
Flow-induced endothelial mitochondrial remodeling mitigates mitochondrial reactive oxygen species production and promotes mitochondrial DNA integrity in a p53-dependent manner.Redox Biol. 2022 Apr;50:102252. doi: 10.1016/j.redox.2022.102252. Epub 2022 Jan 29. Redox Biol. 2022. PMID: 35121402 Free PMC article.
References
-
- Achanta G, Haung P (2004) Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res 64: 6233–6239 - PubMed
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 - PubMed
-
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

