An online database for brain disease research
- PMID: 16594998
- PMCID: PMC1489945
- DOI: 10.1186/1471-2164-7-70
An online database for brain disease research
Abstract
Background: The Stanley Medical Research Institute online genomics database (SMRIDB) is a comprehensive web-based system for understanding the genetic effects of human brain disease (i.e. bipolar, schizophrenia, and depression). This database contains fully annotated clinical metadata and gene expression patterns generated within 12 controlled studies across 6 different microarray platforms.
Description: A thorough collection of gene expression summaries are provided, inclusive of patient demographics, disease subclasses, regulated biological pathways, and functional classifications.
Conclusion: The combination of database content, structure, and query speed offers researchers an efficient tool for data mining of brain disease complete with information such as: cross-platform comparisons, biomarkers elucidation for _target discovery, and lifestyle/demographic associations to brain diseases.
Figures
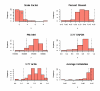



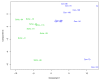


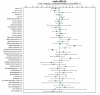
Similar articles
-
MILANO--custom annotation of microarray results using automatic literature searches.BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article.
-
The genopolis microarray database.BMC Bioinformatics. 2007 Mar 8;8 Suppl 1(Suppl 1):S21. doi: 10.1186/1471-2105-8-S1-S21. BMC Bioinformatics. 2007. PMID: 17430566 Free PMC article.
-
p53FamTaG: a database resource of human p53, p63 and p73 direct _target genes combining in silico prediction and microarray data.BMC Bioinformatics. 2007 Mar 8;8 Suppl 1(Suppl 1):S20. doi: 10.1186/1471-2105-8-S1-S20. BMC Bioinformatics. 2007. PMID: 17430565 Free PMC article.
-
Gene expression profiling in schizophrenia and related mental disorders.Neuroscientist. 2006 Aug;12(4):349-61. doi: 10.1177/1073858406287536. Neuroscientist. 2006. PMID: 16840711 Review.
-
Collecting, storing, and mining research data in a brain bank.Handb Clin Neurol. 2018;150:167-179. doi: 10.1016/B978-0-444-63639-3.00013-X. Handb Clin Neurol. 2018. PMID: 29496139 Review.
Cited by
-
A _targeted sequencing study of glutamatergic candidate genes in suicide attempters with bipolar disorder.Am J Med Genet B Neuropsychiatr Genet. 2016 Dec;171(8):1080-1087. doi: 10.1002/ajmg.b.32479. Epub 2016 Aug 2. Am J Med Genet B Neuropsychiatr Genet. 2016. PMID: 27480506 Free PMC article.
-
Involvement of PTPN5, the gene encoding the striatal-enriched protein tyrosine phosphatase, in schizophrenia and cognition.Psychiatr Genet. 2012 Aug;22(4):168-76. doi: 10.1097/YPG.0b013e3283518586. Psychiatr Genet. 2012. PMID: 22555153 Free PMC article.
-
Identification of the gene signature reflecting schizophrenia's etiology by constructing artificial intelligence-based method of enhanced reproducibility.CNS Neurosci Ther. 2019 Sep;25(9):1054-1063. doi: 10.1111/cns.13196. Epub 2019 Jul 27. CNS Neurosci Ther. 2019. PMID: 31350824 Free PMC article.
-
SZGR: a comprehensive schizophrenia gene resource.Mol Psychiatry. 2010 May;15(5):453-62. doi: 10.1038/mp.2009.93. Mol Psychiatry. 2010. PMID: 20424623 Free PMC article.
-
Male-specific association of the 2p25 region with suicide attempt in bipolar disorder.J Psychiatr Res. 2020 Feb;121:151-158. doi: 10.1016/j.jpsychires.2019.11.009. Epub 2019 Nov 18. J Psychiatr Res. 2020. PMID: 31830721 Free PMC article.
References
-
- Iacobas DA, Urban M, Iacobas S, Spray DC. Control and variability of gene expression in mouse brain and in a neuroblastoma cell line. Rom J Physiol. 2003;39–40:2002–71. - PubMed
-
- Jurata LW, Bukhman YV, Charles V, Capriglione F, Bullard J, Lemire AL, Mohammed A, Pham Q, Laeng P, Brockman JA, Altar CA. Comparison of microarray-based mRMA profiling technologies for identification of psychiatric disease and drug signatures. J Neurosci Methods. 2004;138:173–88. doi: 10.1016/j.jneumeth.2004.04.002. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

