The bovine QTL viewer: a web accessible database of bovine Quantitative Trait Loci
- PMID: 16753058
- PMCID: PMC1508159
- DOI: 10.1186/1471-2105-7-283
The bovine QTL viewer: a web accessible database of bovine Quantitative Trait Loci
Abstract
Background: Many important agricultural traits such as weight gain, milk fat content and intramuscular fat (marbling) in cattle are quantitative traits. Most of the information on these traits has not previously been integrated into a genomic context. Without such integration application of these data to agricultural enterprises will remain slow and inefficient. Our goal was to populate a genomic database with data mined from the bovine quantitative trait literature and to make these data available in a genomic context to researchers via a user friendly query interface.
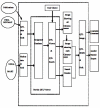
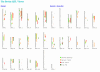
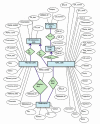
Description: The QTL (Quantitative Trait Locus) data and related information for bovine QTL are gathered from published work and from existing databases. An integrated database schema was designed and the database (MySQL) populated with the gathered data. The bovine QTL Viewer was developed for the integration of QTL data available for cattle. The tool consists of an integrated database of bovine QTL and the QTL viewer to display QTL and their chromosomal position.
Conclusion: We present a web accessible, integrated database of bovine (dairy and beef cattle) QTL for use by animal geneticists. The viewer and database are of general applicability to any livestock species for which there are public QTL data. The viewer can be accessed at http://bovineqtl.tamu.edu.
Figures

Similar articles
-
A gene-based high-resolution comparative radiation hybrid map as a framework for genome sequence assembly of a bovine chromosome 6 region associated with QTL for growth, body composition, and milk performance traits.BMC Genomics. 2006 Mar 16;7:53. doi: 10.1186/1471-2164-7-53. BMC Genomics. 2006. PMID: 16542434 Free PMC article.
-
Bovine quantitative trait loci analysis for growth, carcass, and meat quality traits in an F2 population from a cross between Japanese Black and Limousin.J Anim Sci. 2008 Nov;86(11):2821-32. doi: 10.2527/jas.2007-0676. Epub 2008 Jul 3. J Anim Sci. 2008. PMID: 18599673
-
AnimalQTLdb: a livestock QTL database tool set for positional QTL information mining and beyond.Nucleic Acids Res. 2007 Jan;35(Database issue):D604-9. doi: 10.1093/nar/gkl946. Epub 2006 Nov 29. Nucleic Acids Res. 2007. PMID: 17135205 Free PMC article.
-
Invited review: quantitative trait nucleotide determination in the era of genomic selection.J Dairy Sci. 2011 Mar;94(3):1082-90. doi: 10.3168/jds.2010-3793. J Dairy Sci. 2011. PMID: 21338774 Review.
-
Quantitative trait locus analysis using J/qtl.Methods Mol Biol. 2009;573:175-88. doi: 10.1007/978-1-60761-247-6_10. Methods Mol Biol. 2009. PMID: 19763928 Review.
Cited by
-
Whole genome linkage disequilibrium maps in cattle.BMC Genet. 2007 Oct 25;8:74. doi: 10.1186/1471-2156-8-74. BMC Genet. 2007. PMID: 17961247 Free PMC article.
-
QTL global meta-analysis: are trait determining genes clustered?BMC Genomics. 2009 Apr 24;10:184. doi: 10.1186/1471-2164-10-184. BMC Genomics. 2009. PMID: 19393059 Free PMC article.
-
A physical map of the bovine genome.Genome Biol. 2007;8(8):R165. doi: 10.1186/gb-2007-8-8-r165. Genome Biol. 2007. PMID: 17697342 Free PMC article.
-
A radiation hybrid map of river buffalo (Bubalus bubalis) chromosome 1 (BBU1).Cytogenet Genome Res. 2007;119(1-2):100-4. doi: 10.1159/000109625. Epub 2007 Dec 14. Cytogenet Genome Res. 2007. PMID: 18160788 Free PMC article.
-
A radiation hybrid map of river buffalo (Bubalus bubalis) chromosome 7 and comparative mapping to the cattle and human genomes.Cytogenet Genome Res. 2007;119(3-4):235-41. doi: 10.1159/000112067. Epub 2008 Feb 1. Cytogenet Genome Res. 2007. PMID: 18253035 Free PMC article.
References
-
- Kappes SM, Keele JW, Stone RT, Sonstegard TS, Smith TPL, McGraw RA, LopezCorrales NL, Beattie CW. A second-generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. - PubMed
-
- Weiss B, Piry S, Steinbach D, Samson F, Eggen A. Bovmap database http://locus.jouy.inra.fr/cgi-bin/bovmap/intro2.pl
-
- Khatkar MS, Thomson PC, Tammen I, Costa F, Raadsma HW. Online Combined QTL Map of Dairy Cattle Traits http://www.vetsci.usyd.edu.au/reprogen/QTL_Map/
MeSH terms
LinkOut - more resources
Full Text Sources

