An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity
- PMID: 17283066
- PMCID: PMC1899900
- DOI: 10.1128/MCB.01098-06
An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity
Abstract
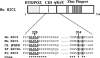
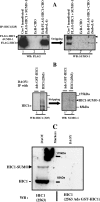
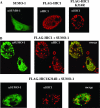
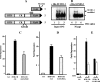
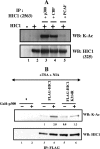
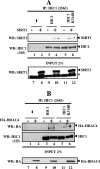
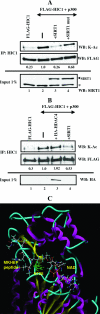
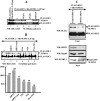
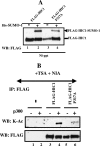
Tumor suppressor HIC1 (hypermethylated in cancer 1) is a gene that is essential for mammalian development, epigenetically silenced in many human tumors, and involved in a complex pathway regulating P53 tumor suppression activity. HIC1 encodes a sequence-specific transcriptional repressor containing five Krüppel-like C(2)H(2) zinc fingers and an N-terminal BTB/POZ repression domain. Here, we show that endogenous HIC1 is SUMOylated in vivo on a phylogenetically conserved lysine, K314, located in the central region which is a second repression domain. K314R mutation does not influence HIC1 subnuclear localization but significantly reduces its transcriptional repression potential, as does the mutation of the other conserved residue in the psiKXE consensus, E316A, or the overexpression of the deSUMOylase SSP3/SENP2. Furthermore, HIC1 is acetylated in vitro by P300/CBP. Strikingly, the K314R mutant is less acetylated than wild-type HIC1, suggesting that this lysine is a _target for both SUMOylation and acetylation. We further show that HIC1 transcriptional repression activity is positively controlled by two types of deacetylases, SIRT1 and HDAC4, which increase the deacetylation and SUMOylation, respectively, of K314. Knockdown of endogenous SIRT1 by the transfection of short interfering RNA causes a significant loss of HIC1 SUMOylation. Thus, this dual-deacetylase complex induces either a phosphorylation-dependent acetylation-SUMOylation switch through a psiKXEXXSP motif, as previously shown for MEF2, or a phosphorylation-independent switch through a psiKXEP motif, as shown here for HIC1, since P317A mutation severely impairs HIC1 acetylation. Finally, our results demonstrate that HIC1 is a _target of the class III deacetylase SIRT1 and identify a new posttranslational modification step in the P53-HIC1-SIRT1 regulatory loop.
Figures

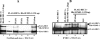

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function.Mol Cell Biol. 2002 May;22(10):3373-88. doi: 10.1128/MCB.22.10.3373-3388.2002. Mol Cell Biol. 2002. PMID: 11971970 Free PMC article.
-
Akt1 sequentially phosphorylates p27kip1 within a conserved but non-canonical region.Cell Div. 2006 Jun 16;1:11. doi: 10.1186/1747-1028-1-11. Cell Div. 2006. PMID: 16780593 Free PMC article.
-
Genedrive kit for detecting single nucleotide polymorphism m.1555A>G in neonates and their mothers: a systematic review and cost-effectiveness analysis.Health Technol Assess. 2024 Oct;28(75):1-75. doi: 10.3310/TGAC4201. Health Technol Assess. 2024. PMID: 39487741 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Ubc9 acetylation: a new route for achieving specificity in substrate SUMOylation.EMBO J. 2013 Mar 20;32(6):773-4. doi: 10.1038/emboj.2013.21. Epub 2013 Feb 8. EMBO J. 2013. PMID: 23395903 Free PMC article.
-
Reduced SIRT1 and SIRT3 and Lower Antioxidant Capacity of Seminal Plasma Is Associated with Shorter Sperm Telomere Length in Oligospermic Men.Int J Mol Sci. 2024 Jan 5;25(2):718. doi: 10.3390/ijms25020718. Int J Mol Sci. 2024. PMID: 38255792 Free PMC article.
-
Strategies to Identify Recognition Signals and _targets of SUMOylation.Biochem Res Int. 2012;2012:875148. doi: 10.1155/2012/875148. Epub 2012 Jul 1. Biochem Res Int. 2012. PMID: 22811915 Free PMC article.
-
Differential regulation of HIC1 _target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells.Mol Cell Biol. 2010 Aug;30(16):4045-59. doi: 10.1128/MCB.00582-09. Epub 2010 Jun 14. Mol Cell Biol. 2010. PMID: 20547755 Free PMC article.
-
ZFP451-mediated SUMOylation of SATB2 drives embryonic stem cell differentiation.Genes Dev. 2021 Aug 1;35(15-16):1142-1160. doi: 10.1101/gad.345843.120. Epub 2021 Jul 8. Genes Dev. 2021. PMID: 34244292 Free PMC article.
References
-
- Bereshchenko, O. R., W. Gu, and R. Dalla-Favera. 2002. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 32:606-613. - PubMed
-
- Bertrand, S., S. Pinte, N. Stankovic-Valentin, S. Deltour-Balerdi, C. Guerardel, A. Begue, V. Laudet, and D. Leprince. 2004. Identification and developmental expression of the zebrafish orthologue of the tumor suppressor gene HIC1. Biochim. Biophys. Acta 1678:57-66. - PubMed
-
- Blander, G., J. Olejnik, E. Krzymanska-Olejnik, T. McDonagh, M. Haigis, M. B. Yaffe, and L. Guarente. 2005. SIRT1 shows no substrate specificity in vitro. J. Biol. Chem. 280:9780-9785. - PubMed
-
- Bossis, G., and F. Melchior. 2006. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21:349-357. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
