Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos
- PMID: 17325035
- PMCID: PMC1899977
- DOI: 10.1128/MCB.00066-07
Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos
Abstract
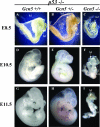
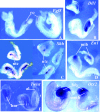
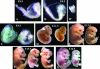
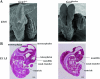
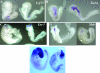
Gcn5 was the first transcription-related histone acetyltransferase (HAT) to be identified. However, the functions of this enzyme in mammalian cells remain poorly defined. Deletion of Gcn5 in mice leads to early embryonic lethality with increased apoptosis in mesodermal lineages. Here we show that deletion of p53 allows Gcn5(-/-) embryos to survive longer, but Gcn5(-/-) p53(-/-) embryos still die in midgestation. Interestingly, embryos homozygous for point mutations in the Gcn5 catalytic domain survive significantly longer than Gcn5(-/-) or Gcn5(-/-) p53(-/-) mice. In contrast to Gcn5(-/-) embryos, Gcn5(hat/hat) embryos do not exhibit increased apoptosis but do exhibit severe cranial neural tube closure defects and exencephaly. Together, our results indicate that Gcn5 has important, HAT-independent functions in early development and that Gcn5 acetyltransferase activity is required for cranial neural tube closure in the mouse.
Figures

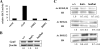


Similar articles
-
Developmental potential of Gcn5(-/-) embryonic stem cells in vivo and in vitro.Dev Dyn. 2007 Jun;236(6):1547-57. doi: 10.1002/dvdy.21160. Dev Dyn. 2007. PMID: 17440986
-
Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos.Dev Dyn. 2008 Apr;237(4):928-40. doi: 10.1002/dvdy.21479. Dev Dyn. 2008. PMID: 18330926 Free PMC article.
-
Proper Gcn5 histone acetyltransferase expression is required for normal anteroposterior patterning of the mouse skeleton.Dev Growth Differ. 2008 Jun;50(5):321-30. doi: 10.1111/j.1440-169X.2008.01041.x. Epub 2008 Apr 22. Dev Growth Differ. 2008. PMID: 18430026 Free PMC article.
-
Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation.Oncogene. 2007 Aug 13;26(37):5341-57. doi: 10.1038/sj.onc.1210604. Oncogene. 2007. PMID: 17694077 Review.
-
Catalysis by protein acetyltransferase Gcn5.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194627. doi: 10.1016/j.bbagrm.2020.194627. Epub 2020 Aug 22. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32841743 Free PMC article. Review.
Cited by
-
Genetics and development of neural tube defects.J Pathol. 2010 Jan;220(2):217-30. doi: 10.1002/path.2643. J Pathol. 2010. PMID: 19918803 Free PMC article. Review.
-
A unique missense allele of BAF155, a core BAF chromatin remodeling complex protein, causes neural tube closure defects in mice.Dev Neurobiol. 2014 May;74(5):483-97. doi: 10.1002/dneu.22142. Epub 2014 Jan 9. Dev Neurobiol. 2014. PMID: 24170322 Free PMC article.
-
GCN5 acetylation is required for craniofacial chondrocyte maturation.Dev Biol. 2020 Aug 1;464(1):24-34. doi: 10.1016/j.ydbio.2020.05.006. Epub 2020 May 22. Dev Biol. 2020. PMID: 32446700 Free PMC article.
-
Epigenetic programming of hypoxic-ischemic encephalopathy in response to fetal hypoxia.Prog Neurobiol. 2015 Jan;124:28-48. doi: 10.1016/j.pneurobio.2014.11.001. Epub 2014 Nov 11. Prog Neurobiol. 2015. PMID: 25450949 Free PMC article. Review.
-
The Histone Acetyltransferase Gcn5 Positively Regulates T Cell Activation.J Immunol. 2017 May 15;198(10):3927-3938. doi: 10.4049/jimmunol.1600312. Epub 2017 Apr 19. J Immunol. 2017. PMID: 28424240 Free PMC article.
References
-
- Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. - PubMed
-
- Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286:2151-2153. - PubMed
-
- Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. - PubMed
-
- Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
