Strategy for encoding and comparison of gene expression signatures
- PMID: 17612401
- PMCID: PMC2323223
- DOI: 10.1186/gb-2007-8-7-r133
Strategy for encoding and comparison of gene expression signatures
Abstract
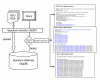
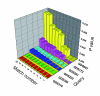
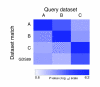
EXALT (EXpression signature AnaLysis Tool) is a computational system enabling comparisons of microarray data across experimental platforms and different laboratories http://seq.mc.vanderbilt.edu/exalt/. An essential feature of EXALT is a database holding thousands of gene expression signatures extracted from the Gene Expression Omnibus, and encoded in a searchable format. This novel approach to performing global comparisons of shared microarray data may have enormous value when coupled directly with a shared data repository.
Figures
Similar articles
-
Web-based interrogation of gene expression signatures using EXALT.BMC Bioinformatics. 2009 Dec 14;10:420. doi: 10.1186/1471-2105-10-420. BMC Bioinformatics. 2009. PMID: 20003458 Free PMC article.
-
DSSTox chemical-index files for exposure-related experiments in ArrayExpress and Gene Expression Omnibus: enabling toxico-chemogenomics data linkages.Bioinformatics. 2009 Mar 1;25(5):692-4. doi: 10.1093/bioinformatics/btp042. Epub 2009 Jan 21. Bioinformatics. 2009. PMID: 19158160
-
A novel gene expression index (GEI) with software support for comparing microarray gene signatures.Gene. 2013 Jan 1;512(1):82-8. doi: 10.1016/j.gene.2012.09.101. Epub 2012 Oct 8. Gene. 2013. PMID: 23059903
-
Functional comparison of microarray data across multiple platforms using the method of percentage of overlapping functions.Methods Mol Biol. 2012;802:123-39. doi: 10.1007/978-1-61779-400-1_9. Methods Mol Biol. 2012. PMID: 22130878
-
Bioinformatics analysis of microarray data.Methods Mol Biol. 2009;573:259-84. doi: 10.1007/978-1-60761-247-6_15. Methods Mol Biol. 2009. PMID: 19763933 Review.
Cited by
-
GEOGLE: context mining tool for the correlation between gene expression and the phenotypic distinction.BMC Bioinformatics. 2009 Aug 25;10:264. doi: 10.1186/1471-2105-10-264. BMC Bioinformatics. 2009. PMID: 19703314 Free PMC article.
-
A data similarity-based strategy for meta-analysis of transcriptional profiles in cancer.PLoS One. 2013;8(1):e54979. doi: 10.1371/journal.pone.0054979. Epub 2013 Jan 29. PLoS One. 2013. PMID: 23383020 Free PMC article.
-
SPARCL1 suppresses metastasis in prostate cancer.Mol Oncol. 2013 Dec;7(6):1019-30. doi: 10.1016/j.molonc.2013.07.008. Epub 2013 Jul 20. Mol Oncol. 2013. PMID: 23916135 Free PMC article.
-
MARQ: an online tool to mine GEO for experiments with similar or opposite gene expression signatures.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W228-32. doi: 10.1093/nar/gkq476. Epub 2010 May 31. Nucleic Acids Res. 2010. PMID: 20513648 Free PMC article.
-
Neonatal Metabolomic Profiles Related to Prenatal Arsenic Exposure.Environ Sci Technol. 2017 Jan 3;51(1):625-633. doi: 10.1021/acs.est.6b04374. Epub 2016 Dec 20. Environ Sci Technol. 2017. PMID: 27997141 Free PMC article.
References
-
- EXALT web server http://seq.mc.vanderbilt.edu/exalt/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

