Conformational instability of the MARK3 UBA domain compromises ubiquitin recognition and promotes interaction with the adjacent kinase domain
- PMID: 17726107
- PMCID: PMC1964837
- DOI: 10.1073/pnas.0703012104
Conformational instability of the MARK3 UBA domain compromises ubiquitin recognition and promotes interaction with the adjacent kinase domain
Abstract
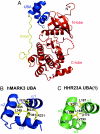
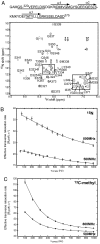
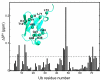
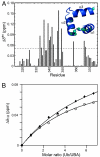
The Par-1/MARK protein kinases play a pivotal role in establishing cellular polarity. This family of kinases contains a unique domain architecture, in which a ubiquitin-associated (UBA) domain is located C-terminal to the kinase domain. We have used a combination of x-ray crystallography and NMR dynamics experiments to understand the interaction of the human (h) MARK3 UBA domain with the adjacent kinase domain as compared with ubiquitin. The x-ray crystal structure of the linked hMARK3 kinase and UBA domains establishes that the UBA domain forms a stable intramolecular interaction with the N-terminal lobe of the kinase domain. However, solution-state NMR studies of the isolated UBA domain indicate that it is highly dynamic, undergoing conformational transitions that can be explained by a folding-unfolding equilibrium. NMR titration experiments indicated that the hMARK3 UBA domain has a detectable but extremely weak affinity for mono ubiquitin, which suggests that conformational instability of the isolated hMARK3 UBA domain attenuates binding to ubiquitin despite the presence of residues typically involved in ubiquitin recognition. Our data identify a molecular mechanism through which the hMARK3 UBA domain has evolved to bind the kinase domain, in a fashion that stabilizes an open conformation of the N- and C-terminal lobes, at the expense of its capacity to engage ubiquitin. These results may be relevant more generally to the 30% of UBA domains that lack significant ubiquitin-binding activity, and they suggest a unique mechanism by which interaction domains may evolve new binding properties.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/Par-1.Structure. 2006 Feb;14(2):173-83. doi: 10.1016/j.str.2005.09.022. Structure. 2006. PMID: 16472737
-
Ubiquitin binding modulates IAP antagonist-stimulated proteasomal degradation of c-IAP1 and c-IAP2(1).Biochem J. 2009 Jan 1;417(1):149-60. doi: 10.1042/BJ20081885. Biochem J. 2009. PMID: 18939944
-
Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions.J Mol Biol. 2002 Jun 21;319(5):1243-55. doi: 10.1016/S0022-2836(02)00302-9. J Mol Biol. 2002. PMID: 12079361
-
Structural and functional studies of mutations affecting the UBA domain of SQSTM1 (p62) which cause Paget's disease of bone.Biochem Soc Trans. 2004 Nov;32(Pt 5):728-30. doi: 10.1042/BST0320728. Biochem Soc Trans. 2004. PMID: 15493999 Review.
-
Microtubule affinity-regulating kinase 4: structure, function, and regulation.Cell Biochem Biophys. 2013 Nov;67(2):485-99. doi: 10.1007/s12013-013-9550-7. Cell Biochem Biophys. 2013. PMID: 23471664 Review.
Cited by
-
Redox Regulation of Brain Selective Kinases BRSK1/2: Implications for Dynamic Control of the Eukaryotic AMPK family through Cys-based mechanisms.bioRxiv [Preprint]. 2024 Apr 10:2023.10.05.561145. doi: 10.1101/2023.10.05.561145. bioRxiv. 2024. PMID: 38586025 Free PMC article. Preprint.
-
Optimization of Selectivity and Pharmacokinetic Properties of Salt-Inducible Kinase Inhibitors that Led to the Discovery of Pan-SIK Inhibitor GLPG3312.J Med Chem. 2024 Jan 11;67(1):380-401. doi: 10.1021/acs.jmedchem.3c01428. Epub 2023 Dec 26. J Med Chem. 2024. PMID: 38147525 Free PMC article.
-
Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity.J Biol Chem. 2010 Nov 12;285(46):36070-80. doi: 10.1074/jbc.M110.145219. Epub 2010 Sep 8. J Biol Chem. 2010. PMID: 20826778 Free PMC article.
-
Loss of Par-1a/MARK3/C-TAK1 kinase leads to reduced adiposity, resistance to hepatic steatosis, and defective gluconeogenesis.Mol Cell Biol. 2010 Nov;30(21):5043-56. doi: 10.1128/MCB.01472-09. Epub 2010 Aug 23. Mol Cell Biol. 2010. PMID: 20733003 Free PMC article.
-
HYPK coordinates degradation of polyneddylated proteins by autophagy.Autophagy. 2022 Aug;18(8):1763-1784. doi: 10.1080/15548627.2021.1997053. Epub 2021 Nov 26. Autophagy. 2022. PMID: 34836490 Free PMC article.
References
-
- Guo S, Kemphues KJ. Cell. 1995;81:611–620. - PubMed
-
- Bohm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Curr Biol. 1997;7:603–606. - PubMed
-
- Shulman JM, Benton R, St Johnston D. Cell. 2000;101:377–388. - PubMed
-
- Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ, III, Wu Z, Stephenson MT, Piwnica-Worms H. Cell Growth Differ. 1998;9:197–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

