The histone methyltransferase SET8 is required for S-phase progression
- PMID: 18166648
- PMCID: PMC2373509
- DOI: 10.1083/jcb.200706150
The histone methyltransferase SET8 is required for S-phase progression
Abstract
Chromatin structure and function is influenced by histone posttranslational modifications. SET8 (also known as PR-Set7 and SETD8) is a histone methyltransferase that monomethylates histonfe H4-K20. However, a function for SET8 in mammalian cell proliferation has not been determined. We show that small interfering RNA inhibition of SET8 expression leads to decreased cell proliferation and accumulation of cells in S phase. This is accompanied by DNA double-strand break (DSB) induction and recruitment of the DNA repair proteins replication protein A, Rad51, and 53BP1 to damaged regions. SET8 depletion causes DNA damage specifically during replication, which induces a Chk1-mediated S-phase checkpoint. Furthermore, we find that SET8 interacts with proliferating cell nuclear antigen through a conserved motif, and SET8 is required for DNA replication fork progression. Finally, codepletion of Rad51, an important homologous recombination repair protein, abrogates the DNA damage after SET8 depletion. Overall, we show that SET8 is essential for genomic stability in mammalian cells and that decreased expression of SET8 results in DNA damage and Chk1-dependent S-phase arrest.
Figures

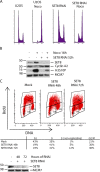
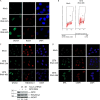

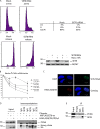
Similar articles
-
PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase.J Cell Biol. 2007 Dec 31;179(7):1413-26. doi: 10.1083/jcb.200706179. Epub 2007 Dec 24. J Cell Biol. 2007. PMID: 18158331 Free PMC article.
-
CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase.Mol Cell. 2010 Oct 8;40(1):22-33. doi: 10.1016/j.molcel.2010.09.015. Mol Cell. 2010. PMID: 20932472 Free PMC article.
-
Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication.J Biol Chem. 2008 Apr 25;283(17):11073-7. doi: 10.1074/jbc.C700242200. Epub 2008 Mar 3. J Biol Chem. 2008. PMID: 18319261 Free PMC article.
-
Roles for the methyltransferase SETD8 in DNA damage repair.Clin Epigenetics. 2022 Mar 4;14(1):34. doi: 10.1186/s13148-022-01251-5. Clin Epigenetics. 2022. PMID: 35246238 Free PMC article. Review.
-
Progress in the Development of Lysine Methyltransferase SETD8 Inhibitors.ChemMedChem. 2016 Aug 19;11(16):1680-5. doi: 10.1002/cmdc.201600272. Epub 2016 Jul 14. ChemMedChem. 2016. PMID: 27411844 Review.
Cited by
-
Prognostic impact of SET domain-containing protein 8 and protein arginine methyltransferase 5 in patients with hepatocellular carcinoma following curative resection.Oncol Lett. 2018 Sep;16(3):3665-3673. doi: 10.3892/ol.2018.9083. Epub 2018 Jul 5. Oncol Lett. 2018. PMID: 30127976 Free PMC article.
-
Biologically active marine natural products and their molecular _targets discovered using a chemical genetics approach.Nat Prod Rep. 2020 May 1;37(5):617-633. doi: 10.1039/c9np00054b. Epub 2019 Nov 21. Nat Prod Rep. 2020. PMID: 31750842 Free PMC article. Review.
-
Lysine methyltransferase inhibitors: where we are now.RSC Chem Biol. 2021 Dec 13;3(4):359-406. doi: 10.1039/d1cb00196e. eCollection 2022 Apr 6. RSC Chem Biol. 2021. PMID: 35441141 Free PMC article. Review.
-
Cell-cycle regulation of non-enzymatic functions of the Drosophila methyltransferase PR-Set7.Nucleic Acids Res. 2018 Apr 6;46(6):2834-2849. doi: 10.1093/nar/gky034. Nucleic Acids Res. 2018. PMID: 29373730 Free PMC article.
-
_targeting the epigenetic machinery of cancer cells.Oncogene. 2015 Jan 8;34(2):135-43. doi: 10.1038/onc.2013.605. Epub 2014 Jan 27. Oncogene. 2015. PMID: 24469033 Review.
References
-
- Bell, S.P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333–374. - PubMed
-
- Fang, J., Q. Feng, C.S. Ketel, H. Wang, R. Cao, L. Xia, H. Erdjument-Bromage, P. Tempst, J.A. Simon, and Y. Zhang. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12:1086–1099. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

