Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation
- PMID: 18172164
- PMCID: PMC2151013
- DOI: 10.1101/gad.1609708
Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation
Abstract
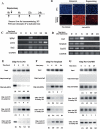
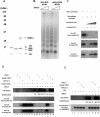
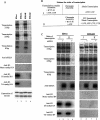
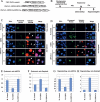
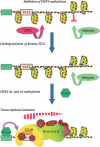
Transcriptional initiation is a key step in the control of mRNA synthesis and is intimately related to chromatin structure and histone modification. Here, we show that the ubiquitylation of H2A (ubH2A) correlates with silent chromatin and regulates transcriptional initiation. The levels of ubH2A vary during hepatocyte regeneration, and based on microarray expression data from regenerating liver, we identified USP21, a ubiquitin-specific protease that catalyzes the hydrolysis of ubH2A. When chromatin is assembled in vitro, ubH2A, but not H2A, specifically represses the di- and trimethylation of H3K4. USP21 relieves this ubH2A-specific repression. In addition, in vitro transcription analysis revealed that ubH2A represses transcriptional initiation, but not transcriptional elongation, by inhibiting H3K4 methylation. Notably, ubH2A-mediated repression was not observed when H3 Lys 4 was changed to arginine. Furthermore, overexpression of USP21 in the liver up-regulates a gene that is normally down-regulated during hepatocyte regeneration. Our studies revealed a novel mode of trans-histone cross-talk, in which H2A ubiquitylation controls the di- and trimethylation of H3K4, resulting in regulation of transcriptional initiation.
Figures

Similar articles
-
The USP21 short variant (USP21SV) lacking NES, located mostly in the nucleus in vivo, activates transcription by deubiquitylating ubH2A in vitro.PLoS One. 2013 Nov 22;8(11):e79813. doi: 10.1371/journal.pone.0079813. eCollection 2013. PLoS One. 2013. PMID: 24278184 Free PMC article.
-
Interaction of the Jhd2 Histone H3 Lys-4 Demethylase with Chromatin Is Controlled by Histone H2A Surfaces and Restricted by H2B Ubiquitination.J Biol Chem. 2015 Nov 27;290(48):28760-77. doi: 10.1074/jbc.M115.693085. Epub 2015 Oct 8. J Biol Chem. 2015. PMID: 26451043 Free PMC article.
-
Histone H2A Ubiquitination Reinforces Mechanical Stability and Asymmetry at the Single-Nucleosome Level.J Am Chem Soc. 2020 Feb 19;142(7):3340-3345. doi: 10.1021/jacs.9b12448. Epub 2020 Feb 4. J Am Chem Soc. 2020. PMID: 32003988
-
Core histone H2A ubiquitylation and transcriptional regulation.Exp Cell Res. 2010 Oct 15;316(17):2707-12. doi: 10.1016/j.yexcr.2010.05.028. Epub 2010 May 31. Exp Cell Res. 2010. PMID: 20685273 Review.
-
Histone H2A ubiquitination in transcriptional regulation and DNA damage repair.Int J Biochem Cell Biol. 2009 Jan;41(1):12-5. doi: 10.1016/j.biocel.2008.09.016. Epub 2008 Sep 26. Int J Biochem Cell Biol. 2009. PMID: 18929679 Review.
Cited by
-
Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis.Int J Mol Sci. 2022 Oct 14;23(20):12285. doi: 10.3390/ijms232012285. Int J Mol Sci. 2022. PMID: 36293135 Free PMC article. Review.
-
Protein monoubiquitylation: _targets and diverse functions.Genes Cells. 2015 Jul;20(7):543-62. doi: 10.1111/gtc.12250. Epub 2015 Jun 18. Genes Cells. 2015. PMID: 26085183 Free PMC article. Review.
-
DUBbing Cancer: Deubiquitylating Enzymes Involved in Epigenetics, DNA Damage and the Cell Cycle As Therapeutic _targets.Front Genet. 2016 Jul 28;7:133. doi: 10.3389/fgene.2016.00133. eCollection 2016. Front Genet. 2016. PMID: 27516771 Free PMC article. Review.
-
Deubiquitination and Stabilization of PD-L1 by USP21.Am J Transl Res. 2021 Nov 15;13(11):12763-12774. eCollection 2021. Am J Transl Res. 2021. PMID: 34956491 Free PMC article.
-
Enzymatic assays for assessing histone deubiquitylation activity.Methods. 2011 Jul;54(3):339-47. doi: 10.1016/j.ymeth.2011.04.001. Epub 2011 Apr 12. Methods. 2011. PMID: 21513801 Free PMC article.
References
-
- Bannister A.J., Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. - PubMed
-
- Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. - PubMed
-
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
