Suppression subtractive hybridization profiles of radial growth phase and metastatic melanoma cell lines reveal novel potential _targets
- PMID: 18211678
- PMCID: PMC2267200
- DOI: 10.1186/1471-2407-8-19
Suppression subtractive hybridization profiles of radial growth phase and metastatic melanoma cell lines reveal novel potential _targets
Abstract
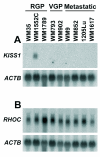
Background: Melanoma progression occurs through three major stages: radial growth phase (RGP), confined to the epidermis; vertical growth phase (VGP), when the tumor has invaded into the dermis; and metastasis. In this work, we used suppression subtractive hybridization (SSH) to investigate the molecular signature of melanoma progression, by comparing a group of metastatic cell lines with an RGP-like cell line showing characteristics of early neoplastic lesions including expression of the metastasis suppressor KISS1, lack of alphavbeta3-integrin and low levels of RHOC.
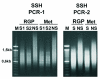
Methods: Two subtracted cDNA collections were obtained, one (RGP library) by subtracting the RGP cell line (WM1552C) cDNA from a cDNA pool from four metastatic cell lines (WM9, WM852, 1205Lu and WM1617), and the other (Met library) by the reverse subtraction. Clones were sequenced and annotated, and expression validation was done by Northern blot and RT-PCR. Gene Ontology annotation and searches in large-scale melanoma expression studies were done for the genes identified.
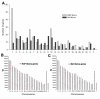
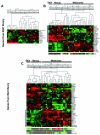
Results: We identified 367 clones from the RGP library and 386 from the Met library, of which 351 and 368, respectively, match human mRNA sequences, representing 288 and 217 annotated genes. We confirmed the differential expression of all genes selected for validation. In the Met library, we found an enrichment of genes in the growth factors/receptor, adhesion and motility categories whereas in the RGP library, enriched categories were nucleotide biosynthesis, DNA packing/repair, and macromolecular/vesicular trafficking. Interestingly, 19% of the genes from the RGP library map to chromosome 1 against 4% of the ones from Met library.
Conclusion: This study identifies two populations of genes differentially expressed between melanoma cell lines from two tumor stages and suggests that these sets of genes represent profiles of less aggressive versus metastatic melanomas. A search for expression profiles of melanoma in available expression study databases allowed us to point to a great potential of involvement in tumor progression for several of the genes identified here. A few sequences obtained here may also contribute to extend annotated mRNAs or to the identification of novel transcripts.
Figures

Similar articles
-
Osteonectin/SPARC induction by ectopic beta(3) integrin in human radial growth phase primary melanoma cells.Cancer Res. 2002 Jan 1;62(1):226-32. Cancer Res. 2002. PMID: 11782382
-
Screening and identification of lung cancer metastasis-related genes by suppression subtractive hybridization.Thorac Cancer. 2012 Aug;3(3):207-216. doi: 10.1111/j.1759-7714.2011.00092.x. Thorac Cancer. 2012. PMID: 28920308
-
Antigenic profile of tumor progression stages in human melanocytic nevi and melanomas.Cancer Res. 1989 Sep 15;49(18):5091-6. Cancer Res. 1989. PMID: 2548711
-
Transcriptional regulation of metastasis-related genes in human melanoma.Clin Exp Metastasis. 2003;20(3):251-63. doi: 10.1023/a:1022991302172. Clin Exp Metastasis. 2003. PMID: 12741683 Review.
-
Gene regulation in melanoma progression by the AP-2 transcription factor.Pigment Cell Res. 2001 Apr;14(2):78-85. doi: 10.1034/j.1600-0749.2001.140202.x. Pigment Cell Res. 2001. PMID: 11310795 Review.
Cited by
-
Oligo- and polymetastatic progression in lung metastasis(es) patients is associated with specific microRNAs.PLoS One. 2012;7(12):e50141. doi: 10.1371/journal.pone.0050141. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251360 Free PMC article.
-
Antitumor immunity and cancer stem cells.Ann N Y Acad Sci. 2009 Sep;1176:154-69. doi: 10.1111/j.1749-6632.2009.04568.x. Ann N Y Acad Sci. 2009. PMID: 19796244 Free PMC article.
-
Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma.Sci Rep. 2018 Sep 21;8(1):14190. doi: 10.1038/s41598-018-31170-6. Sci Rep. 2018. PMID: 30242167 Free PMC article.
-
Melanoma stem cells and metastasis: mimicking hematopoietic cell trafficking?Lab Invest. 2014 Jan;94(1):13-30. doi: 10.1038/labinvest.2013.116. Epub 2013 Oct 14. Lab Invest. 2014. PMID: 24126889 Free PMC article. Review.
-
Modulation of T-cell activation by malignant melanoma initiating cells.Cancer Res. 2010 Jan 15;70(2):697-708. doi: 10.1158/0008-5472.CAN-09-1592. Epub 2010 Jan 12. Cancer Res. 2010. PMID: 20068175 Free PMC article.
References
-
- Grossman D, Altieri DC. Drug resistance in melanoma: mechanisms, apoptosis, and new potential therapeutic _targets. Cancer Metastasis Rev. 2001;20:3–11. - PubMed
-
- Helmbach H, Rossmann E, Kern MA, Schadendorf D. Drug-resistance in human melanoma. Int J Cancer. 2001;93:617–622. - PubMed
-
- Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–3151. - PubMed
-
- Meier F, Satyamoorthy K, Nesbit M, Hsu MY, Schittek B, Garbe C, Herlyn M. Molecular events in melanoma development and progression. Front Biosci. 1998;3:D1005–10. - PubMed
-
- Kath R, Jambrosic JA, Holland L, Rodeck U, Herlyn M. Development of invasive and growth factor-independent cell variants from primary human melanomas. Cancer Res. 1991;51:2205–2211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

