Co-localization of the amyloid precursor protein and Notch intracellular domains in nuclear transcription factories
- PMID: 18403052
- PMCID: PMC2868363
- DOI: 10.1016/j.neurobiolaging.2008.03.001
Co-localization of the amyloid precursor protein and Notch intracellular domains in nuclear transcription factories
Abstract
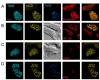
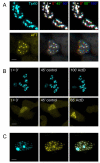
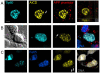
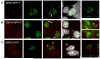
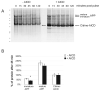
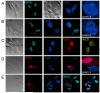
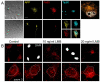
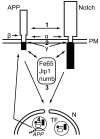
The beta-amyloid precursor protein (APP) plays a major role in Alzheimer's disease. The APP intracellular domain (AICD), together with Fe65 and Tip60, localizes to spherical nuclear AFT complexes, which may represent sites of transcription. Despite a lack of co-localization with several described nuclear compartments, we have identified a close apposition between AFT complexes and splicing speckles, Cajal bodies and PML bodies. Live imaging revealed that AFT complexes were highly mobile within nuclei and following pharmacological inhibition of transcription fused into larger assemblies. We have previously shown that AICD regulates the expression of its own precursor APP. In support of our earlier findings, transfection of APP promoter plasmids as substrates resulted in cytosolic AFT complex formation at labeled APP promoter plasmids. In addition, identification of chromosomal APP or KAI1 gene loci by fluorescence in situ hybridization showed their close association with nuclear AFT complexes. The transcriptional activator Notch intracellular domain (NICD) localized to the same nuclear spots as occupied by AFT complexes suggesting that these nuclear compartments correspond to transcription factories. Fe65 and Tip60 also co-localized with APP in the neurites of primary neurons. Pre-assembled AFT complexes may serve to assist fast nuclear signaling upon endoproteolytic APP cleavage.
Figures


Similar articles
-
Visualization and quantification of APP intracellular domain-mediated nuclear signaling by bimolecular fluorescence complementation.PLoS One. 2013 Sep 25;8(9):e76094. doi: 10.1371/journal.pone.0076094. eCollection 2013. PLoS One. 2013. PMID: 24086696 Free PMC article.
-
A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein.J Biol Chem. 2005 Nov 4;280(44):36895-904. doi: 10.1074/jbc.M502861200. Epub 2005 Aug 15. J Biol Chem. 2005. PMID: 16103124 Free PMC article.
-
The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor.J Cell Sci. 2004 Sep 1;117(Pt 19):4435-48. doi: 10.1242/jcs.01323. J Cell Sci. 2004. PMID: 15331662
-
A putative role of the Amyloid Precursor Protein Intracellular Domain (AICD) in transcription.Acta Neurobiol Exp (Wars). 2008;68(2):219-28. doi: 10.55782/ane-2008-1691. Acta Neurobiol Exp (Wars). 2008. PMID: 18511958 Review.
-
Fe65 matters: new light on an old molecule.IUBMB Life. 2012 Dec;64(12):936-42. doi: 10.1002/iub.1094. Epub 2012 Nov 5. IUBMB Life. 2012. PMID: 23129269 Review.
Cited by
-
Amyloid Precursor Protein (APP) Regulates Gliogenesis and Neurogenesis of Human Neural Stem Cells by Several Signaling Pathways.Int J Mol Sci. 2023 Aug 19;24(16):12964. doi: 10.3390/ijms241612964. Int J Mol Sci. 2023. PMID: 37629148 Free PMC article.
-
Physiological effects of amyloid precursor protein and its derivatives on neural stem cell biology and signaling pathways involved.Neural Regen Res. 2019 Oct;14(10):1661-1671. doi: 10.4103/1673-5374.257511. Neural Regen Res. 2019. PMID: 31169172 Free PMC article.
-
Critical role of presenilin-dependent γ-secretase activity in DNA damage-induced promyelocytic leukemia protein expression and apoptosis.Cell Death Differ. 2013 Apr;20(4):639-48. doi: 10.1038/cdd.2012.162. Epub 2013 Jan 11. Cell Death Differ. 2013. PMID: 23306558 Free PMC article.
-
PuF, an antimetastatic and developmental signaling protein, interacts with the Alzheimer's amyloid-β precursor protein via a tissue-specific proximal regulatory element (PRE).BMC Genomics. 2013 Jan 31;14:68. doi: 10.1186/1471-2164-14-68. BMC Genomics. 2013. PMID: 23368879 Free PMC article.
-
The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway.J Biol Chem. 2010 Dec 31;285(53):41443-54. doi: 10.1074/jbc.M110.141390. Epub 2010 Oct 20. J Biol Chem. 2010. PMID: 20961856 Free PMC article.
References
-
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. - PubMed
-
- Borg JP, Yang Y, De Taddeo-Borg M, Margolis B, Turner RS. The X11alpha protein slows cellular amyloid precursor protein processing and reduces Abeta40 and Abeta42 secretion. J Biol Chem. 1998;273:14761–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

