Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation
- PMID: 18539900
- PMCID: PMC2515143
- DOI: 10.1182/blood-2008-02-137398
Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation
Abstract
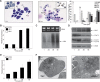
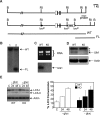
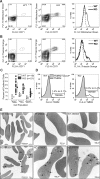
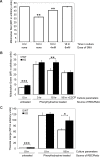
Production of a red blood cell's hemoglobin depends on mitochondrial heme synthesis. However, mature red blood cells are devoid of mitochondria and rely on glycolysis for ATP production. The molecular basis for the selective elimination of mitochondria from mature red blood cells remains controversial. Recent evidence suggests that clearance of both mitochondria and ribosomes, which occurs in reticulocytes following nuclear extrusion, depends on autophagy. Here, we demonstrate that Ulk1, a serine threonine kinase with homology to yeast atg1p, is a critical regulator of mitochondrial and ribosomal clearance during the final stages of erythroid maturation. However, in contrast to the core autophagy genes such as atg5 and atg7, expression of ulk1 is not essential for induction of macroautophagy in response to nutrient deprivation or for survival of newborn mice. Together, these data suggest that the ATG1 homologue, Ulk1, is a component of the selective autophagy machinery that leads to the elimination of organelles in erythroid cells rather that an essential mechanistic component of autophagy.
Figures
Similar articles
-
Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes.Nat Commun. 2014 Jun 4;5:4004. doi: 10.1038/ncomms5004. Nat Commun. 2014. PMID: 24895007
-
A novel role for nuclear factor-erythroid 2 in erythroid maturation by modulation of mitochondrial autophagy.Haematologica. 2016 Sep;101(9):1054-64. doi: 10.3324/haematol.2015.132589. Epub 2016 Jun 16. Haematologica. 2016. PMID: 27479815 Free PMC article.
-
Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation.Blood. 2009 Jul 2;114(1):157-64. doi: 10.1182/blood-2008-04-151639. Epub 2009 May 5. Blood. 2009. PMID: 19417210 Free PMC article.
-
Normal and disordered reticulocyte maturation.Curr Opin Hematol. 2011 May;18(3):152-7. doi: 10.1097/MOH.0b013e328345213e. Curr Opin Hematol. 2011. PMID: 21423015 Free PMC article. Review.
-
Mitochondrial clearance by autophagy in developing erythrocytes: clearly important, but just how much so?Cell Cycle. 2010 May 15;9(10):1901-6. doi: 10.4161/cc.9.10.11603. Epub 2010 May 15. Cell Cycle. 2010. PMID: 20495377 Review.
Cited by
-
SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages.Autophagy. 2016 Aug 2;12(8):1272-91. doi: 10.1080/15548627.2016.1183081. Epub 2016 Jun 23. Autophagy. 2016. PMID: 27337507 Free PMC article.
-
The Interplay of HIV and Autophagy in Early Infection.Front Microbiol. 2021 Apr 28;12:661446. doi: 10.3389/fmicb.2021.661446. eCollection 2021. Front Microbiol. 2021. PMID: 33995324 Free PMC article. Review.
-
Danon disease - dysregulation of autophagy in a multisystem disorder with cardiomyopathy.J Cell Sci. 2016 Jun 1;129(11):2135-43. doi: 10.1242/jcs.184770. Epub 2016 May 10. J Cell Sci. 2016. PMID: 27165304 Free PMC article. Review.
-
A short linear motif in BNIP3L (NIX) mediates mitochondrial clearance in reticulocytes.Autophagy. 2012 Sep;8(9):1325-32. doi: 10.4161/auto.20764. Epub 2012 Aug 21. Autophagy. 2012. PMID: 22906961 Free PMC article.
-
Autophagy and mechanisms of effective immunity.Front Immunol. 2012 Apr 4;3:60. doi: 10.3389/fimmu.2012.00060. eCollection 2012. Front Immunol. 2012. PMID: 22566941 Free PMC article.
References
-
- Geminard C, de Gassart A, Vidal M. Reticulocyte maturation: mitoptosis and exosome release. Biocell. 2002;26:205–215. - PubMed
-
- Pieczenik SR, Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp Mol Pathol. 2007;83:84–92. - PubMed
-
- Schewe T, Rapoport SM, Kuhn H. Enzymology and physiology of reticulocyte lipoxygenase: comparison with other lipoxygenases. Adv Enzymol Relat Areas Mol Biol. 1986;58:191–272. - PubMed
-
- van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. A function for lipoxygenase in programmed organelle degradation. Nature. 1998;395:392–395. - PubMed
-
- Vijayvergiya C, De Angelis D, Walther M, et al. High-level expression of rabbit 15-lipoxygenase induces collapse of the mitochondrial pH gradient in cell culture. Biochemistry. 2004;43:15296–15302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

