A mouse to human search for plasma proteome changes associated with pancreatic tumor development
- PMID: 18547137
- PMCID: PMC2504036
- DOI: 10.1371/journal.pmed.0050123
A mouse to human search for plasma proteome changes associated with pancreatic tumor development
Abstract
Background: The complexity and heterogeneity of the human plasma proteome have presented significant challenges in the identification of protein changes associated with tumor development. Refined genetically engineered mouse (GEM) models of human cancer have been shown to faithfully recapitulate the molecular, biological, and clinical features of human disease. Here, we sought to exploit the merits of a well-characterized GEM model of pancreatic cancer to determine whether proteomics technologies allow identification of protein changes associated with tumor development and whether such changes are relevant to human pancreatic cancer.
Methods and findings: Plasma was sampled from mice at early and advanced stages of tumor development and from matched controls. Using a proteomic approach based on extensive protein fractionation, we confidently identified 1,442 proteins that were distributed across seven orders of magnitude of abundance in plasma. Analysis of proteins chosen on the basis of increased levels in plasma from tumor-bearing mice and corroborating protein or RNA expression in tissue documented concordance in the blood from 30 newly diagnosed patients with pancreatic cancer relative to 30 control specimens. A panel of five proteins selected on the basis of their increased level at an early stage of tumor development in the mouse was tested in a blinded study in 26 humans from the CARET (Carotene and Retinol Efficacy Trial) cohort. The panel discriminated pancreatic cancer cases from matched controls in blood specimens obtained between 7 and 13 mo prior to the development of symptoms and clinical diagnosis of pancreatic cancer.
Conclusions: Our findings indicate that GEM models of cancer, in combination with in-depth proteomic analysis, provide a useful strategy to identify candidate markers applicable to human cancer with potential utility for early detection.
Conflict of interest statement
Figures

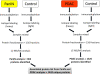
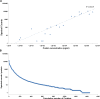

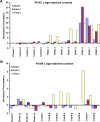
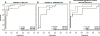
Similar articles
-
Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling.J Proteome Res. 2011 May 6;10(5):2359-76. doi: 10.1021/pr101148r. Epub 2011 Mar 28. J Proteome Res. 2011. PMID: 21443201 Free PMC article.
-
Optomechanical devices for deep plasma cancer proteomics.Semin Cancer Biol. 2018 Oct;52(Pt 1):26-38. doi: 10.1016/j.semcancer.2017.08.011. Epub 2017 Sep 1. Semin Cancer Biol. 2018. PMID: 28867489 Review.
-
Integrated proteomic analysis of human cancer cells and plasma from tumor bearing mice for ovarian cancer biomarker discovery.PLoS One. 2009 Nov 19;4(11):e7916. doi: 10.1371/journal.pone.0007916. PLoS One. 2009. PMID: 19936259 Free PMC article.
-
Proteomic profiling of pancreatic cancer for biomarker discovery.Mol Cell Proteomics. 2005 Apr;4(4):523-33. doi: 10.1074/mcp.R500004-MCP200. Epub 2005 Jan 31. Mol Cell Proteomics. 2005. PMID: 15684406 Review.
-
Identification of novel early pancreatic cancer biomarkers KIF5B and SFRP2 from "first contact" interactions in the tumor microenvironment.J Exp Clin Cancer Res. 2022 Aug 24;41(1):258. doi: 10.1186/s13046-022-02425-y. J Exp Clin Cancer Res. 2022. PMID: 36002889 Free PMC article.
Cited by
-
Multiplex _targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study.J Proteome Res. 2012 Mar 2;11(3):1937-48. doi: 10.1021/pr201117w. Epub 2012 Feb 8. J Proteome Res. 2012. PMID: 22316387 Free PMC article.
-
Perspectives of _targeted mass spectrometry for protein biomarker verification.Curr Opin Chem Biol. 2009 Dec;13(5-6):518-25. doi: 10.1016/j.cbpa.2009.09.014. Epub 2009 Oct 7. Curr Opin Chem Biol. 2009. PMID: 19818677 Free PMC article. Review.
-
Postmenopausal estrogen and progestin effects on the serum proteome.Genome Med. 2009 Dec 24;1(12):121. doi: 10.1186/gm121. Genome Med. 2009. PMID: 20034393 Free PMC article.
-
Pathogenesis of pancreatic cancer: lessons from animal models.Toxicol Pathol. 2014 Jan;42(1):217-28. doi: 10.1177/0192623313508250. Epub 2013 Oct 31. Toxicol Pathol. 2014. PMID: 24178582 Free PMC article. Review.
-
Validation of four candidate pancreatic cancer serological biomarkers that improve the performance of CA19.9.BMC Cancer. 2013 Sep 3;13:404. doi: 10.1186/1471-2407-13-404. BMC Cancer. 2013. PMID: 24007603 Free PMC article.
References
-
- Hanash S. Disease proteomics. Nature. 2003;422:226–232. - PubMed
-
- Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. - PubMed
-
- Faca V, Pitteri SJ, Newcomb L, Glukhova V, Phanstiel D, et al. Contribution of protein fractionation to depth of analysis of the serum and plasma proteomes. J Proteome Res. 2007;6:3558–3565. - PubMed
-
- States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. - PubMed
-
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

