Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19-9 levels in pancreatic cancer
- PMID: 18615426
- PMCID: PMC2992687
- DOI: 10.1002/pmic.200700951
Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19-9 levels in pancreatic cancer
Abstract
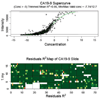
The current study analyzed reverse phase protein arrays (RPPA) as a means to experimentally validate biomarkers in blood samples. One microliter samples of sera (n = 71), and plasma (n = 78) were serially diluted and printed on NC-coated slides. CA19-9 levels from RPPA results were compared with identical patient samples as measured by ELISA. There was a strong correlation between RPPA and ELISA (r = 0.87) as determined by scatter plots. Sample reproducibility of CA19-9 levels was excellent (interslide correlation r = 0.88; intraslide correlation r = 0.83). The ability of RPPA to accurately distinguish CA19-9 levels between cancer and noncancer samples were determined using receiver operating characteristic curves and compared with ELISA. The AUC for RPPA and ELISA was comparable (0.87 and 0.86, respectively). When the mean CA19-9 levels of normal samples was used as a cutoff for RPPA and compared with the standard clinical ELISA cutoff, comparable specificities (71% for both) were observed. Notably, RPPA samples normalized to albumin showed increased sensitivity compared to ELISA (90% vs. 75%). As RPPA is a high-throughput method that shows results comparable to that of ELISA, we propose that RPPA is a viable technique for rapid experimental screening and validation of candidate biomarkers in blood samples.
Conflict of interest statement
The authors declare no financial or commercial conflicts of interest.
Figures






Similar articles
-
Validation of four candidate pancreatic cancer serological biomarkers that improve the performance of CA19.9.BMC Cancer. 2013 Sep 3;13:404. doi: 10.1186/1471-2407-13-404. BMC Cancer. 2013. PMID: 24007603 Free PMC article.
-
Periostin and CA242 as potential diagnostic serum biomarkers complementing CA19.9 in detecting pancreatic cancer.Cancer Sci. 2018 Sep;109(9):2841-2851. doi: 10.1111/cas.13712. Epub 2018 Jul 24. Cancer Sci. 2018. PMID: 29945294 Free PMC article.
-
Trefoil factor(s) and CA19.9: A promising panel for early detection of pancreatic cancer.EBioMedicine. 2019 Apr;42:375-385. doi: 10.1016/j.ebiom.2019.03.056. Epub 2019 Apr 5. EBioMedicine. 2019. PMID: 30956167 Free PMC article.
-
Reverse phase protein arrays in signaling pathways: a data integration perspective.Drug Des Devel Ther. 2015 Jul 7;9:3519-27. doi: 10.2147/DDDT.S38375. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26185419 Free PMC article. Review.
-
Serum carbohydrate antigen 19-9 in pancreatic adenocarcinoma: a mini review for surgeons.ANZ J Surg. 2017 Dec;87(12):987-992. doi: 10.1111/ans.14131. Epub 2017 Aug 13. ANZ J Surg. 2017. PMID: 28803454 Review.
Cited by
-
Reverse phase protein arrays in acute leukemia: investigative and methodological challenges.Expert Rev Proteomics. 2021 Dec;18(12):1087-1097. doi: 10.1080/14789450.2021.2020655. Epub 2021 Dec 29. Expert Rev Proteomics. 2021. PMID: 34965151 Free PMC article. Review.
-
MCU controls melanoma progression through a redox-controlled phenotype switch.EMBO Rep. 2022 Nov 7;23(11):e54746. doi: 10.15252/embr.202254746. Epub 2022 Sep 26. EMBO Rep. 2022. PMID: 36156348 Free PMC article.
-
Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response.Sci Adv. 2020 Oct 2;6(40):eaba5714. doi: 10.1126/sciadv.aba5714. Print 2020 Oct. Sci Adv. 2020. PMID: 33008904 Free PMC article.
-
Increasing the sensitivity of reverse phase protein arrays by antibody-mediated signal amplification.Proteome Sci. 2010 Jun 22;8:36. doi: 10.1186/1477-5956-8-36. Proteome Sci. 2010. PMID: 20569466 Free PMC article.
-
The current state of proteomics in GI oncology.Dig Dis Sci. 2009 Mar;54(3):431-57. doi: 10.1007/s10620-008-0656-5. Epub 2008 Dec 23. Dig Dis Sci. 2009. PMID: 19104933 Free PMC article. Review.
References
-
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol.Cell Proteomics. 2002;1:845–867. - PubMed
-
- Etzioni R, Urban N, Ramsey S, McIntosh M, et al. The case for early detection. Nat. Rev. Cancer. 2003;3:243–252. - PubMed
-
- Grote T, Logsdon CD. Progress on molecular markers of pancreatic cancer. Curr. Opin. Gastroenterol. 2007;23:508–514. - PubMed
-
- Jemal A, Siegel R, Ward E, Murray T, et al. Cancer statistics. 2007 CA Cancer J. Clin. 2007;57:43–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

